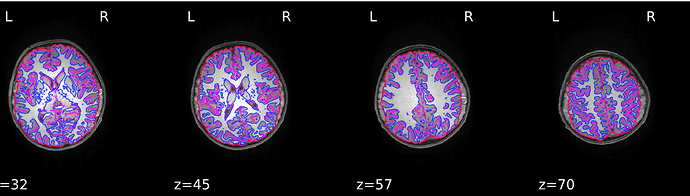
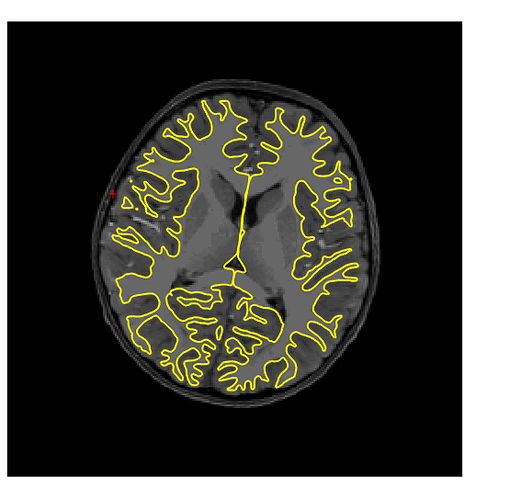
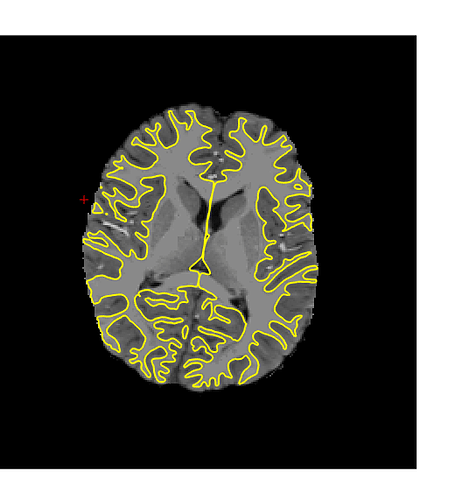
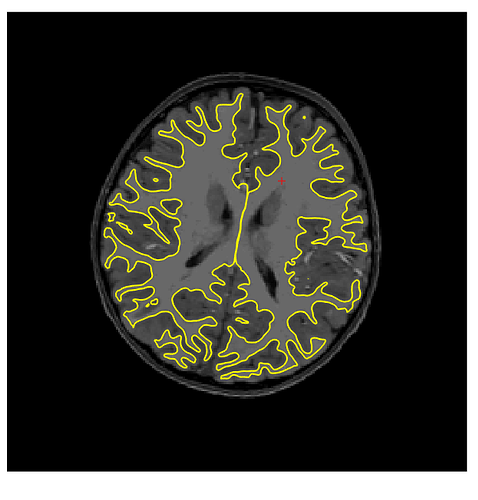
Hmmm. Sorry again for the delay, I just got back to lab after two months of travel. I am still getting the same type of error with templateflow (below), running with 1.4.1rc4. I think this might be because our singularity installation doesn’t have a /scratch dir in the container. Not sure why, I assumed that was bundled with the container? Am I missing something? Some googling yielded that probably file system overlay is not enable on my Linux cluster, or the particular singularity configuration is different…?
Running the command above yields:
WARNING: Skipping user bind, non existent bind point (directory) in container: '/scratch'
TEMPLATEFLOW_HOME=/scratch/templateflow
The error:
Running fMRIPREP version 1.4.1rc2:
* BIDS dataset path: /mnt.
* Participant list: ['CBPD0142'].
* Run identifier: 20190716-191532_434bd2ab-c16e-46da-96d2-5ba907b1ce6d.
Downloading https://templateflow.s3.amazonaws.com/tpl-MNIPediatricAsym/cohort-2/tpl-MNIPediatricAsym_cohort-2_res-1_T1w.nii.gz
Process Process-2:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/multiprocessing/process.py", line 297, in _bootstrap
self.run()
File "/usr/local/miniconda/lib/python3.7/multiprocessing/process.py", line 99, in run
self._target(*self._args, **self._kwargs)
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/cli/run.py", line 608, in build_workflow
work_dir=str(work_dir),
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/base.py", line 259, in init_fmriprep_wf
use_syn=use_syn,
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/base.py", line 551, in init_single_subject_wf
skull_strip_template=skull_strip_template,
File "/usr/local/miniconda/lib/python3.7/site-packages/smriprep/workflows/anatomical.py", line 230, in init_anat_preproc_wf
normalization_quality='precise' if not debug else 'testing')
File "/usr/local/miniconda/lib/python3.7/site-packages/niworkflows/anat/ants.py", line 188, in init_brain_extraction_wf
tpl_target_path = get_template(in_template, **template_spec)
File "/usr/local/miniconda/lib/python3.7/site-packages/templateflow/api.py", line 39, in get
_s3_get(filepath)
File "/usr/local/miniconda/lib/python3.7/site-packages/templateflow/api.py", line 139, in _s3_get
with filepath.open('wb') as f:
File "/usr/local/miniconda/lib/python3.7/pathlib.py", line 1176, in open
opener=self._opener)
File "/usr/local/miniconda/lib/python3.7/pathlib.py", line 1030, in _opener
return self._accessor.open(self, flags, mode)
OSError: [Errno 30] Read-only file system: '/opt/templateflow/tpl-MNIPediatricAsym/cohort-2/tpl-MNIPediatricAsym_cohort-2_res-1_T1w.nii.gz'