Hello, I’ve recently preprocessed some fMRI data with fMRIPrep and I was interested in having the data in a surface space, i.e. a mesh, so I used the --output-spaces fsnative command for that.
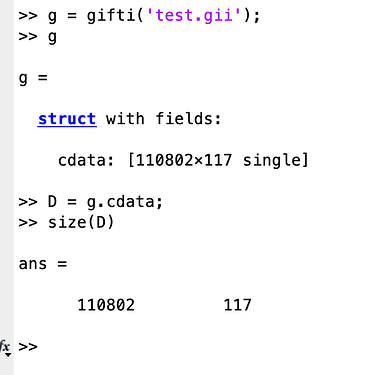
I now have the output data and I wanted to have a look at some specific regions but since my BOLD signal data looks like this:
As a vertices x timepoints array (in particular, 110802 vertices x 117 timepoints). Now, I’m not sure how to proceed.
By the way, this code was done with MATLAB and I used this Github library to open the .gii file.
Is there any default brain atlas for brain parcellation in fMRIPrep? Or is there any other way I can extract the areas from this array?
Thanks a lot,
Arnau.
 ).
).