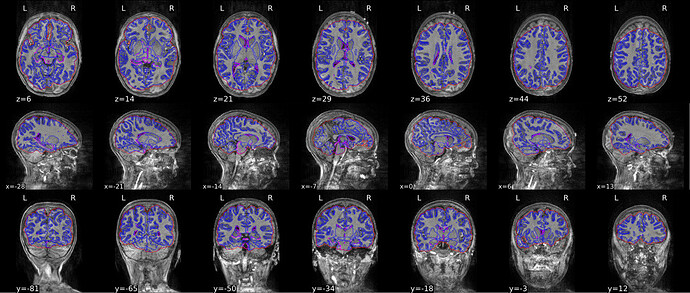
I have a subject with rather poor quality T1w anatomical data. This is causing the brain mask to go awry and this error then propagates down the rest of the fmriprep pipeline. I have searched the forum, but I haven’t figured out the recommended way to try to patch up the brain mask to salvage this subject’s data. I was thinking about running BETS or some other brain extraction software on the T1w volume before running fmriprep and then setting the --skull-strip-t1w flag to skip. Any thoughts about whether this is the best workaround, or if it will have the desired effect?