Hi all,
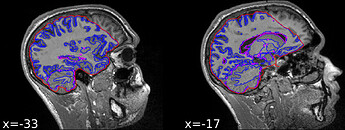
I have a T1-weighted MPRAGE image that I’m using as an anatomical reference for my BOLD scans of a young adult participant. Running fMRIprep on one of my subjects, I encountered an issue with a suboptimal brain mask:
Notably, it seems like this subject has more signal than others around the mouth.
I was hoping someone could advise me how I could manually intervene (or change the behavior of antsBrainExtraction.sh) to obtain a better mask.
Would it be best to skull-strip the raw T1w image and use the “skip” option for --skull-strip-t1w flag as suggested by @dittothat here?
Many thanks,
Roey
P.S.
Looking at the surface reconstruction on top of the participant’s T1w template reveals that FreeSurfer also find it hard to deal with this image as well:
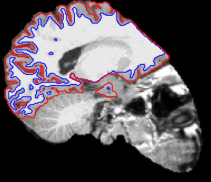
Thanks for tagging me in your question. I never did get to the bottom of this issue. If I figure it out, I will let you know.
Yes, manually skull-stripping and then skipping skull-stripping in fMRIPrep would work, and is probably the easiest solution.
To fix this for others, though, I would first verify that running antsBrainExtraction.sh directly produces the same issue. If it doesn’t, then we need to figure out what’s happening differently. If it does, you could possibly work with the ANTs team to come up with a more robust pipeline that we could incorporate.
Thanks, @effigies!
Semi-manual skull-stripping (first with FSL’s bet and then manual corrections of the mask using MRtrix’s mrview) did the trick. After running fMRIPrep on this skull-stripped T1w everything seems to work fine:
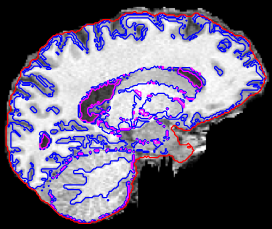
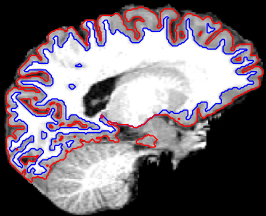
Including FreeSurfer:
Thanks for the quick advice.
Best,
Roey
We saw a lot of skull strip issues in our data. Usually to do with the raw T1w data having really strong non-uniformities. @jerdra ended up doing a bunch of extra non-uniformity pre-fmriprep to compensate.
@jerdra knows the whole situation better than I.