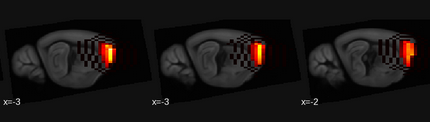
Hello,
I’m seeking assistance with a persistent issue I’m encountering while performing ICA on mouse brain rs-fMRI data. Despite my efforts, checkerboard artifacts continue to appear around the components, and I’m unsure of the best approach to eliminate these artifacts.
For context, I’ve employed a standard preprocessing pipeline that includes slice timing correction, bias correction, motion correction, spike sorting, segmentation, normalization to the Allen Institute atlas, and initial smoothing with a 4x3x2 voxel kernel (I intentionally applied this initial smoothing to minimize the need for further smoothing during ICA), filtering in the range [0.01; 0.22].
I’ve used the CanICA algorithm from nilearn with the following parameters:
canica = CanICA(mask=mask, n_components=Ncomponent, memory="nilearn_cache", memory_level=2, verbose=0, random_state=0, detrend=False, threshold='auto', standardize=True, n_init=10, low_pass=None, high_pass=None, mask_strategy="background", n_jobs=1)
I would greatly appreciate any suggestions!