Summary of what happened:
Hello,
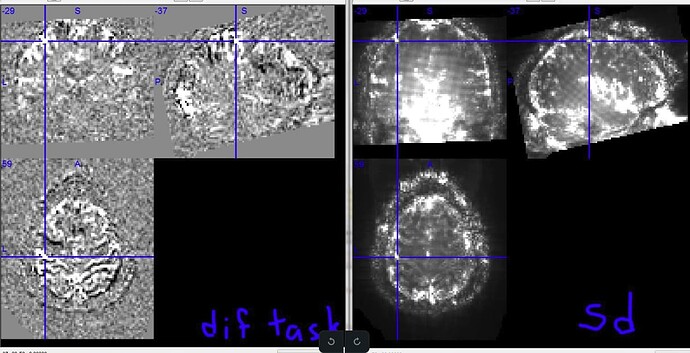
I am trying to select a sequence for a new experiment at 7T. We have two options: a single echo (SE) sequence and a multiecho sequence (ME). The SE has 1.2 mm isotropic voxels and the ME has 1.5mm isotropic voxels. I have preprocessed the raw images with fmriprep in native, T1w, MNI152NLin6Asym-res1, and MNI152NLin6Asym-res2. The images themselves look ok, but I am noticing a strange checkerboard pattern (most evident in the coronal view) in their tSNR:
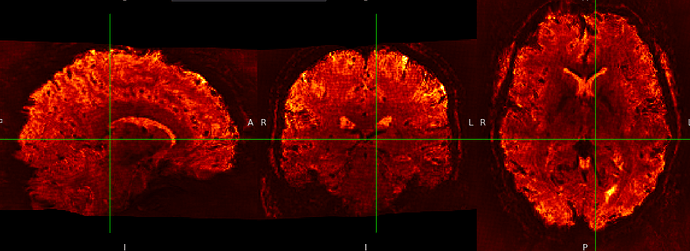
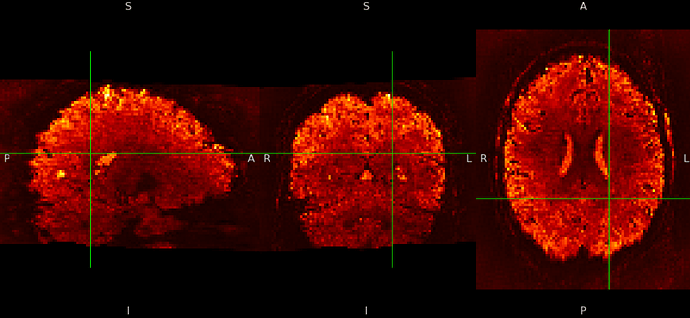
SE sequence in MNI152NLin6Asym-res1
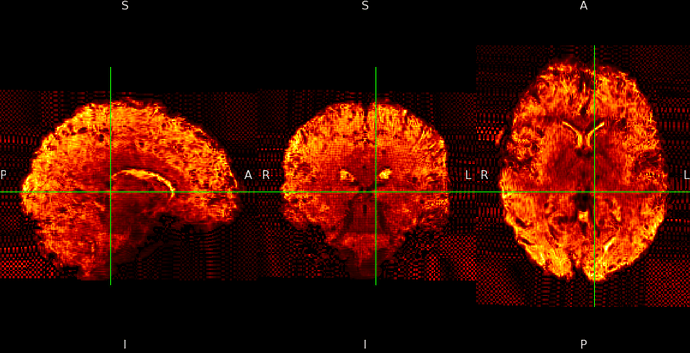
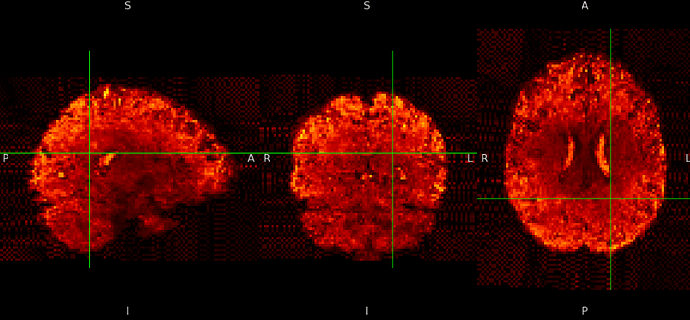
ME sequence in MNI152NLin6Asym-res1
The checkerboard pattern is more evident in the ME sequence where it is also visible outside the brain, but it is present in both, ME and SE inside the brain.
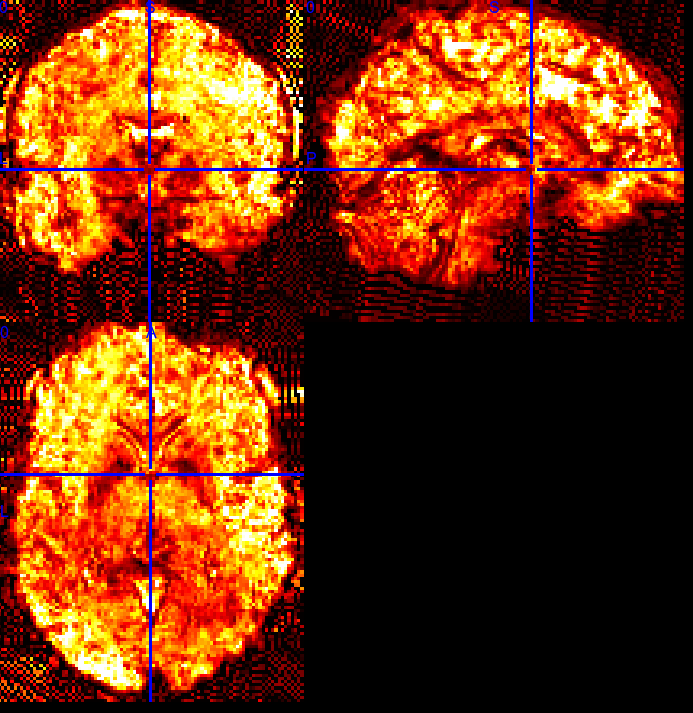
The pattern is not present in the raw data. It is not present either in the fmriprep native space. For example,
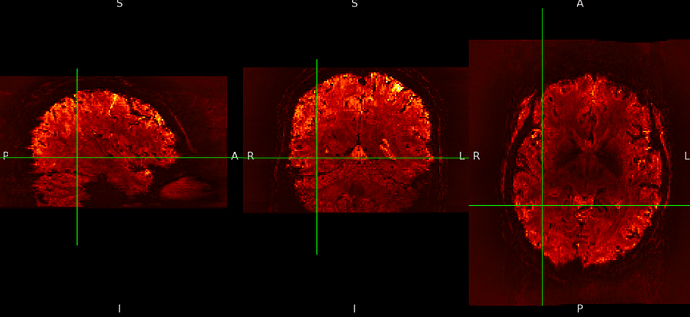
SE sequence in native:
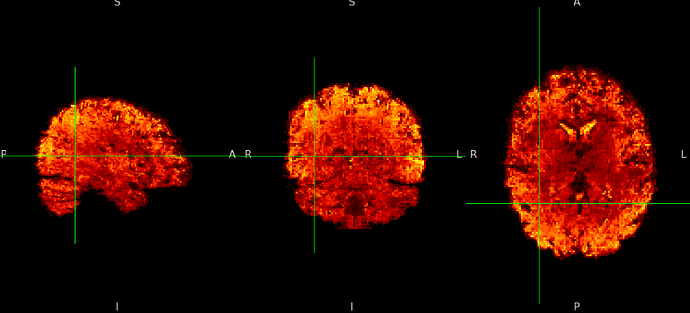
ME sequence in native:
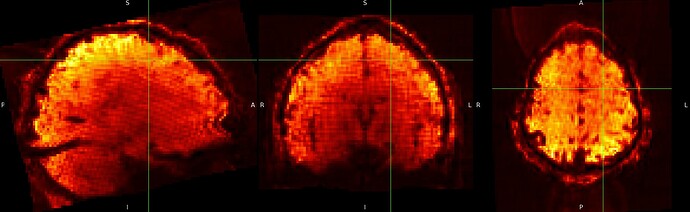
And although it is not visible for the SE sequence in MNI152NLin6Asym-res2, it is visible outside the brain in the ME sequence in MNI152NLin6Asym-res2
SE sequence in MNI152NLin6Asym-res2
ME sequence in MNI152NLin6Asym-res2
I am here puzzled over:
- why is this pattern visible only at MNI 1mm resolutions? Please remember that my voxels are between 1.2 and 1.5 mm isotropic.
- Could this be related to the Lanczos interpolation?
- why is the pattern exacerbated in the ME sequence? Is it the echo combination?
- What could be done to solve this?
Thank you so much!
-Ana
Commands used (and if a helper script was used, a link to the helper script or the command generated):
- to compute tsNR:
fslmaths bold.nii.gz -Tmean bold_mean.nii.gz
fslmaths bold.nii.gz -Tstd bold_std.nii.gz
fslmaths bold_mean.nii.gz -div bold_std.nii.gz TSNR_map.nii.gz
- for fMRIprep:
singularity run --cleanenv \
-B $SINGULARITYENV_TEMPLATEFLOW_HOME:/home/.cache/templateflow \
./singularity_images/fmriprep-24.1.1.simg \
${BIDS_DIR} \
${BIDS_DIR}/${DERIVS_DIR} \
participant --participant-label ${subject} \
-w ${WORK_DIR} \
--output-spaces MNI152NLin6Asym:res-1:res-2 T1w func \
--ignore slicetiming \
--fs-no-reconall \
--fs-license-file ./singularity_images/freesurfer_license.txt \
--fd-spike-threshold 0.2 \
--notrack \
Version:
fmriprep . 24.1.1
Environment (Docker, Singularity / Apptainer, custom installation):
singularity
Data formatted according to a validatable standard?
The data is in BIDS format and was validated before running the command using the online BIDS validator. The command I used also forced the validation and there are no errors reported in the html. Is this enough?
Unfortunately, I deleted all working directories and I do not know where to get this info. How can I access it?
Relevant log outputs (up to 20 lines):
The command ran successfully… so I am unsure what is needed here. Could you be more specific of what should I be looking for?
Screenshots / relevant information:
nipype_version = “1.8.6”
templateflow_version = “24.2.0”
fmriprep version = “24.1.1”
fsl version 6.0.7