As a newcomer to fMRI analysis and algorithms, I have recently tried to use Nilearn to apply Searchlight-SVR techniques to mining the relationship between resting data(fALFF) and behavioral scale scores. I don’t know how to implement it at the moment, the example in official documentation is very clear, but it’s a comparison of task states, I don’t know how to modify it to apply to my research, hope to get help!
Some details are required:
1)the searchlight_score uses the coefficient of correlation between the predicted value and the true value.
2)the .nii file of mapping, with the purpose of subsequently obtaining the corresponding brain region.
Please clarify what inputs you are using and what association you want to compute in the Searchlight analysis.
Best,
Bertrand
The input is a 4-D file(.nii); it is a merge file of 504 people’s 3-D data. And, another input is 504x1 scale scores(.csv). what i want compute in the searchlight is the correlation coefficient between y_predict and y_true by SVR.
One of the problems I encountered was, (as the code shown in the figure), how to set the “scoring=” in nilearn.decoding.SearchLight() to the coefficient of correlation between the predicted value and the true value?
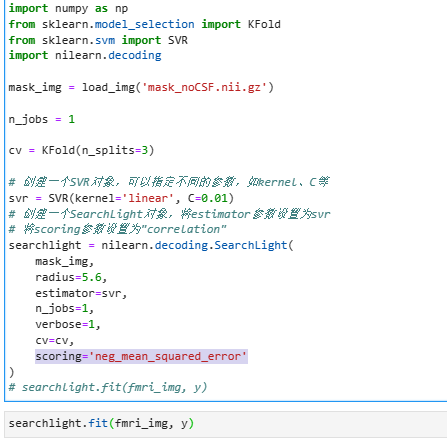
The admissible metrics are here: 3.3. Metrics and scoring: quantifying the quality of predictions — scikit-learn 1.3.1 documentation
Indeed, i don’t see correlation.
You can take a look at
3.3.1.3. Implementing your own scoring object¶
Does this help ?
Best,
Bertrand
I know these are the allowed indicators, I mean can I customize this parameter? For example, I want to output correlations (true vs. predicted)
