Thanks so much for your help, @dowdlelt.
Could you please tell me how I might set the number of components directly? I could not find documentation of such an option on TEDANA’s manual.
Thanks!
Roey
Of course, should have mentioned it directly - its the --tedpca option, detailed here, relevant bit:
–tedpca
Method with which to select components in TEDPCA. PCA decomposition with the mdl, kic and aic options is based on a Moving Average (stationary Gaussian) process and are ordered from most to least aggressive. ‘kundu’ or ‘kundu-stabilize’ are selection methods that were distributed with MEICA. Users may also provide a float from 0 to 1, in which case components will be selected based on the cumulative variance explained or an integer greater than 1 in which case the specificed number of components will be selected.
You could also specify variance explained with a float. Unfortunately, guidance kind of breaks down at this point, it is hard to recommend any specific settings - we often think about 1/3rd of the timepoints works well, but its all so dataset dependent!
I’ve seen the shared data, I’ll try and take a look.
Thanks for pointing me to the --tedpca option! I missed the integer part there.
Unfortunately this didn’t help in terms of getting cleaner data.
I did upload the TEDANA reports for the additional data (in the same Drive folder). As you predicted, using FLEET doesn’t seem to make a strong difference (but may have helped with tSNR as you said). There it seems like there is a strong global signal that is not braing cleaned well by TEDANA.
MB4_GRP2 (curvefit+aic)
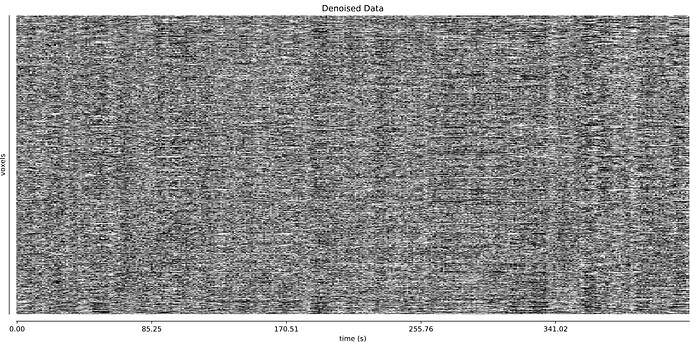
optimally combined + denoised:
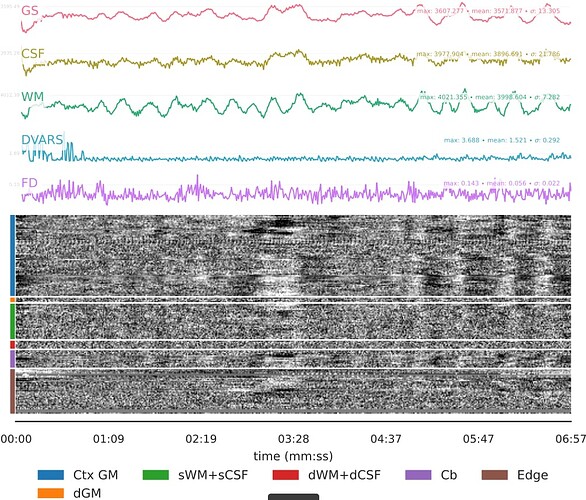
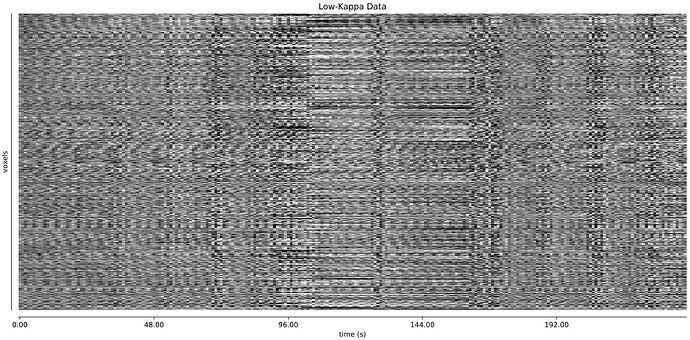
I’m alittle worried about this second dataset. Perhaps these large flctuations are respiratory, and hard for TEDANA to handle? See these fMRIprep QC plots:
These large fluctuations might stem from the subject closing their eyes every once in a while, since overlaying the BOLD signal from some voxels in the eye shows clar correspondence between the GS (red) and the eye signal (blue), shown in another scan of that subject:
Thanks again for all your help!
Roey
Returning to Kundu et al., (2013), I’m surprised that a voxel size of 3.2 mm would be an issue, as they used voxels of 3.75 mm.
I wasn’t very clear there - Less about the specific voxel size, but maybe the high level of acceleration. The PCA/ICA steps depend a lot on the noise levels of the data - so you have high SNR, but also a fair amount of noise from acceleration plus extra cross voxel correlations due to that same acceleration.
I see. Thanks!
I will try scanning a full Fourier protocol to reduce the voxels correlations.
Do you think despiking the data after HMC and SDC but before tedana is also worth a try ?
I suppose I wasn’t clear enough - I wasn’t intending that to be a recommendation, full fourier is going to take much more time (in the TE dimension), and I’m not sure you could spare it. I think 7/8ths is fine. It may be worth adding an additional echo onto the faster one - which would sacrifice a bit more time, but may give you better signal to work with. 3 echoes is necessary, but 4, 5, 6 etc can be helpful depending on the situation.
I’m still planning on taking a closer look at what you have shared - just wanted to make this note quickly.
Hi NeuroStarsers, especially @dowdlelt!
I would greatly appreciate your insights regarding the latest data I’ve collected. Here I chose to reduce the MB factor from 4 to 3, collecting full Fourier data (at the expanse of a longer TR of 1093 ms).
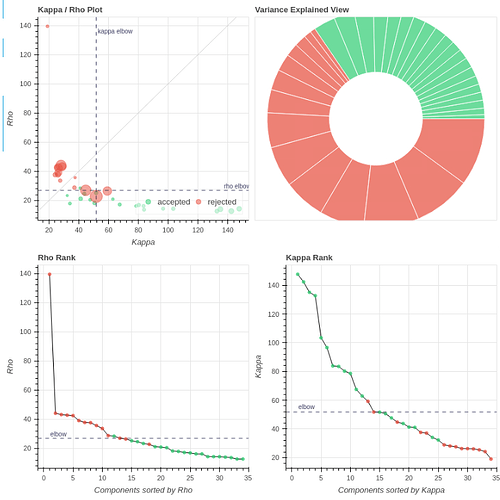
The tedana QC plots (full reports are here on Drive):
You can see that a larger percentage of the variance is epxlained by accepted components, although the number of components is still surprisingly low (35 components for ~400 volumes).
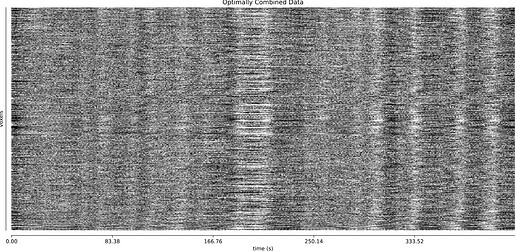
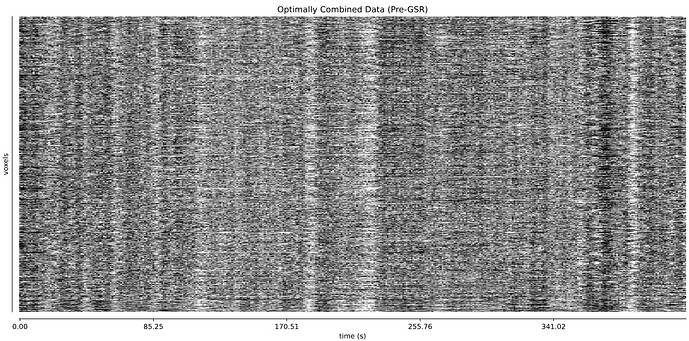
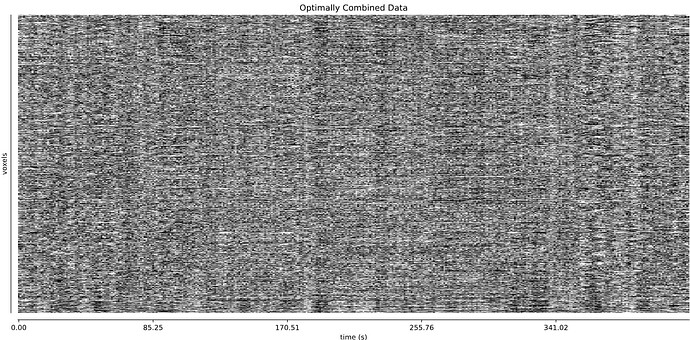
The thing I’m worried about is the large amount of global signal artifacts, here in the optimally combined data:
GSR really gets rid of most of it:
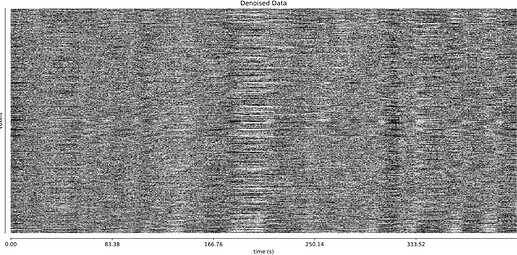
And it’s a little hard to tell whether TE-dependent denoising makes a large difference:
I would love to hear what you think about it.
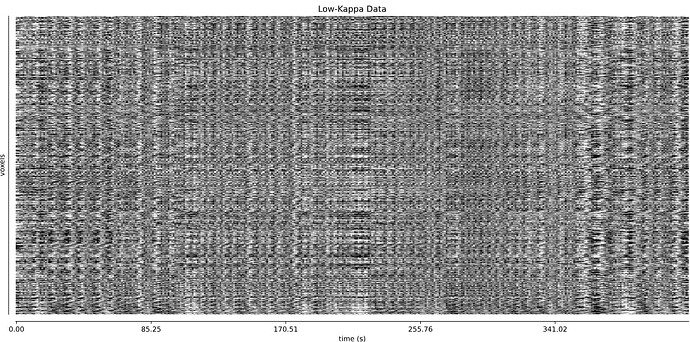
Also, since I don’t have much experience in tedana, I’m not sure if the following signal for the rejected components looks reasonable; I’m worried that the repeated patterns hereare indicative of some underlying problem I’m missing:
As always, your help is greatly appreciated!
Enjoy the weekend ![]()
I’m still chewing on this - sorry for all the back and forth, and the bit of confusion, but the data is surprising me, in that its not behaving as I would expect, and nor is tedana.
Is there a reason to prefer fast sampling? I’m a fan personally, but I tend to think in 7T terms these days where things are a bit different. As I think I said before, maybe it is worth dragging out those echo times - with more echoes (to be clear, even if you need 7/8ths partial fourier - its not evil, technically).
I’m thinking in particular of the Lynch paper - https://www.sciencedirect.com/science/article/pii/S2211124720315291?via%3Dihub#sec4
From their methods:
Multi-echo, multi-band resting-state fMRI scans were collected using a T2∗-weighted echo-planar sequence covering the full brain (TR: 1355 ms; TE1: 13.40 ms, TE2: 31.11 ms, TE3: 48.82 ms, TE4: 66.53 ms, and TE5: 84.24 ms; FOV: 216 mm; flip angle: 68°; 2.4mm isotropic; 72 slices; AP phase encoding direction; in-plane acceleration factor: 2; and multi-band acceleration factor: 6) with 640 volumes acquired per scan for a total acquisition time of 14 min and 27 s.
I’m not sure that you would need to match exactly those parameters (perhaps stick with MB4, no need to go to 6!) - your relaxed voxel size may allow for less acceleration for example. I’m mostly recommending this because I don’t feel like the combined data looks quite like I would expect. I think those longer echoes could matter a lot. Some part of me recalls that the last echo should be approximately 2 x T2*, ie about ~66 or so? This could explain why your “slower” protocol had somewhat better behavior.
I think if you take your “faster” protocol, and add a few more echoes, out towards 70ms, you may see much better performance. @handwerkerd tends to be a bit smarter about this, so I’m curious what he thinks. I’m just getting the feeling from your data that the faster sampling is only hurtning you - and with 5 echoes (or so?), and a TR of 1~1.3 seconds you’d see very nice signal with that voxel size.
Thank you again for these insights, @dowdlelt!
As for the sampling, we do need a TR or around 1s for the purpose of our experiment.
I wasn’t aware of this 2 x T2* rule-of-thumb. Thanks for bringing it to my attention.
I downloaded some online ME datasets to get a better feeling of what I might expect in our own data. This included the Multi-Echo Cambridge dataset with
TE: 12, 28, 44, 60 ms
TR = 2.47
GRAPPA 3 (I think not MB at all)
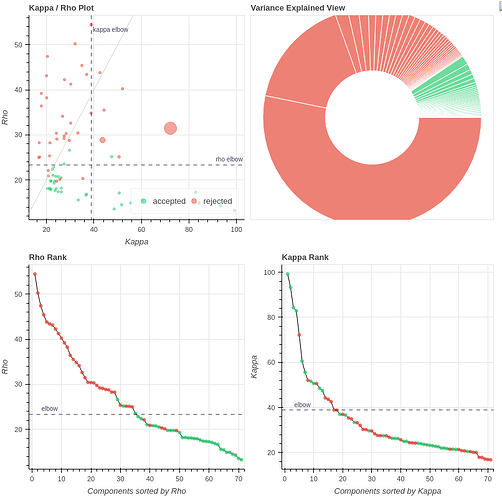
I ran fmriprep (this time with slice-timing correction due to the slow TR) and ran tedana with either all 4 echos or only the first 3 (which is more similar to our own data). I found the results quite perplexing. For example, for one subject (04570):
Tedana, curvefit, AIC, no GSR, 3 echos
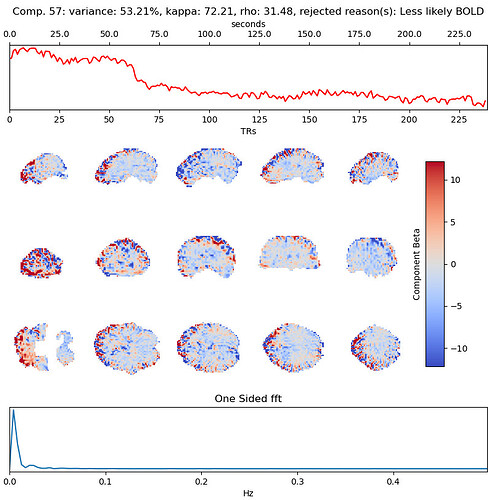
with the first excluded component being (53% variance explained):
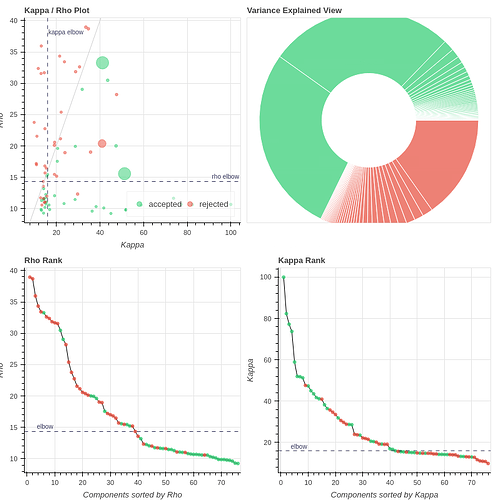
Tedana, curvefit, AIC, no GSR, 4 echos
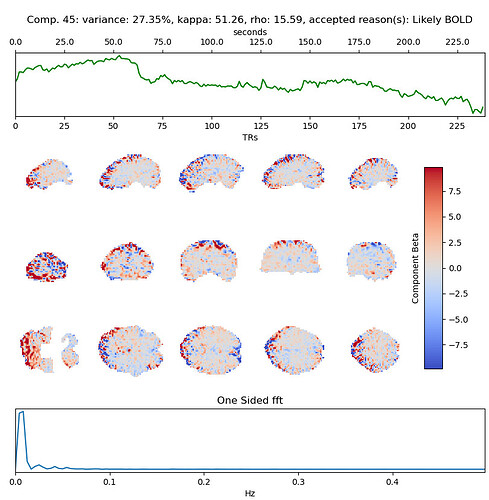
with the 2nd accepted component (27% variance explained) being quite similar to the excluded one when we use only 3 echos:
Indeed, there are many more ICs compared with our data (~75) with only 139 volumes per scan.
I’m confused by this extreme difference in tedana’s behavior when taking 3 or 4 echos.
I will also add that for some subjects (like 11310), the carpet plot for the rejected components does have a similar repetitive texture to it, like we see in our data:
Thanks for staying with me here ![]()
A quick note because I found the manuscript I was thinking of - Theoretical optimization of multi-echo fMRI data acquisition
If my quick read is right, then the target is ~1.5 x T2* of the tissue, so ~60 is perhaps even on the low end. In other words, the more echoes, the better, and the longer the better (up to a point).
I think the analysis you’ve done there in instructive for this - that last echo is adding a lot of useful information about the BOLD contrast, and therefor helping the fit. I’ll try and take a closer look shortly.
Thanks, Logan!
I also uploaded the reports (and figures folders) as a fourth dataset here.
Thank you for the reference! I will look into it.
with the 2nd accepted component (27% variance explained) being quite similar to the excluded one when we use only 3 echos:
So you think that this large component, with this specific timecourse is in fact BOLD related and should be rtained? My intuition was that this is indeed a bad component that should be discarded, but perhaps I’m wrong. Looking at the denoised data, the 3-echo version actually seems cleaner to me (number of echos appears in the top left label):
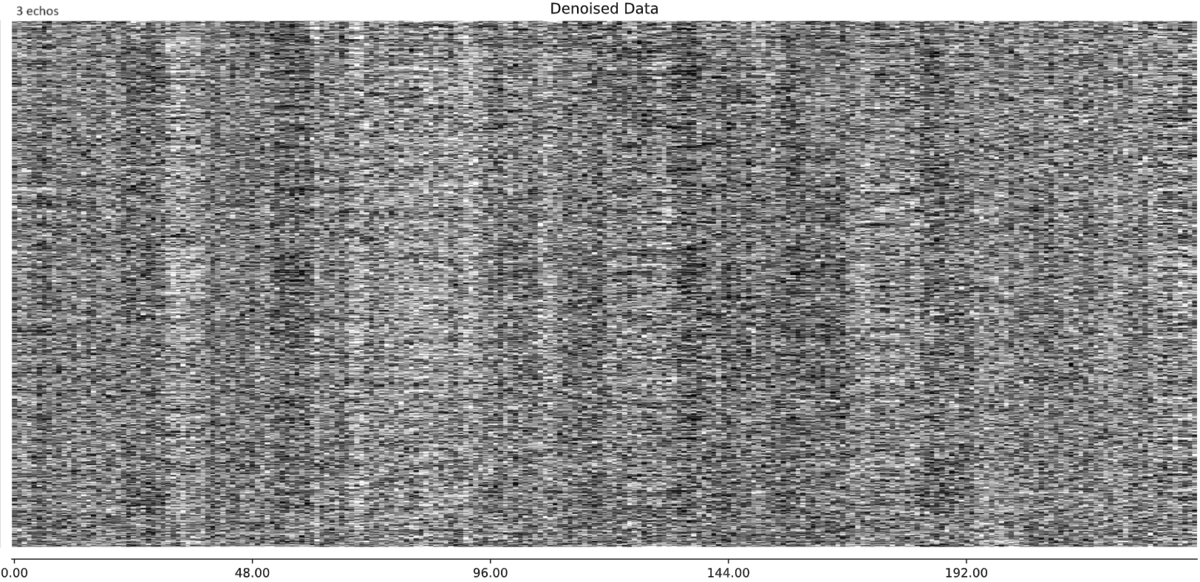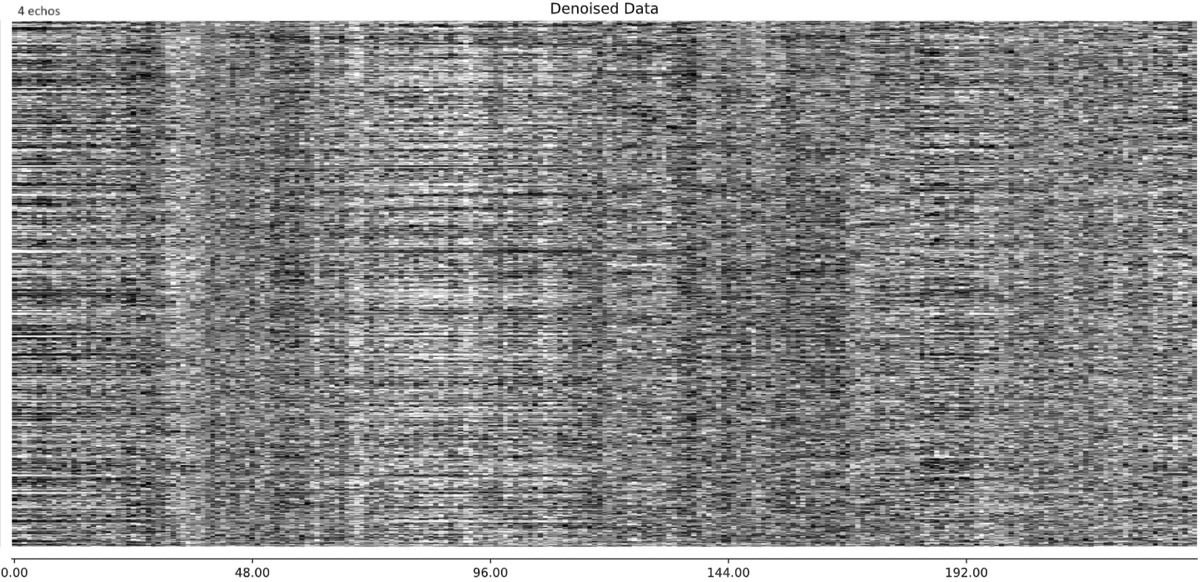
and similarly for another subject:
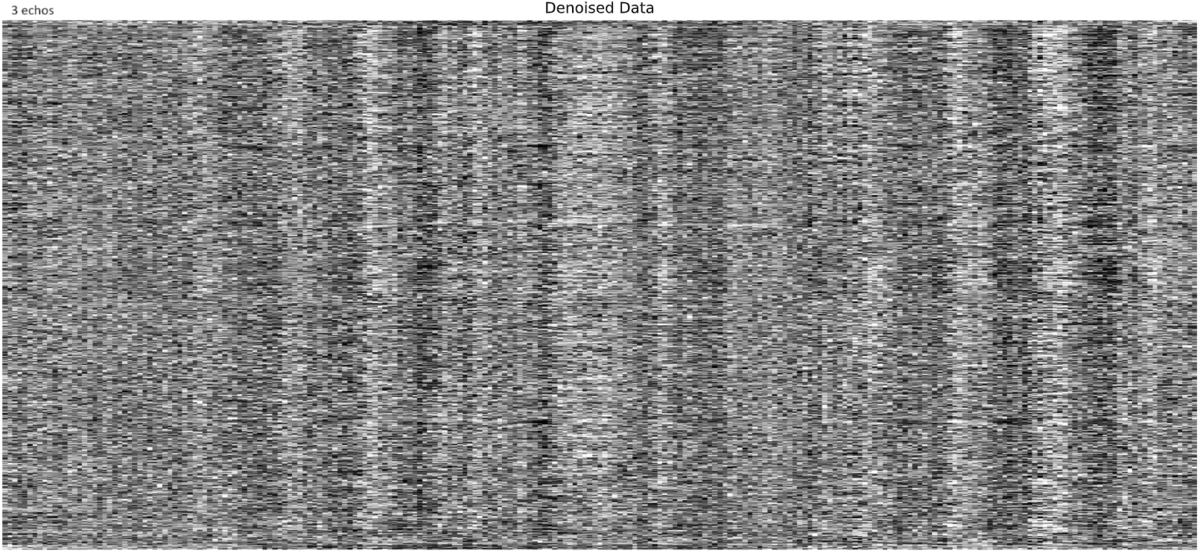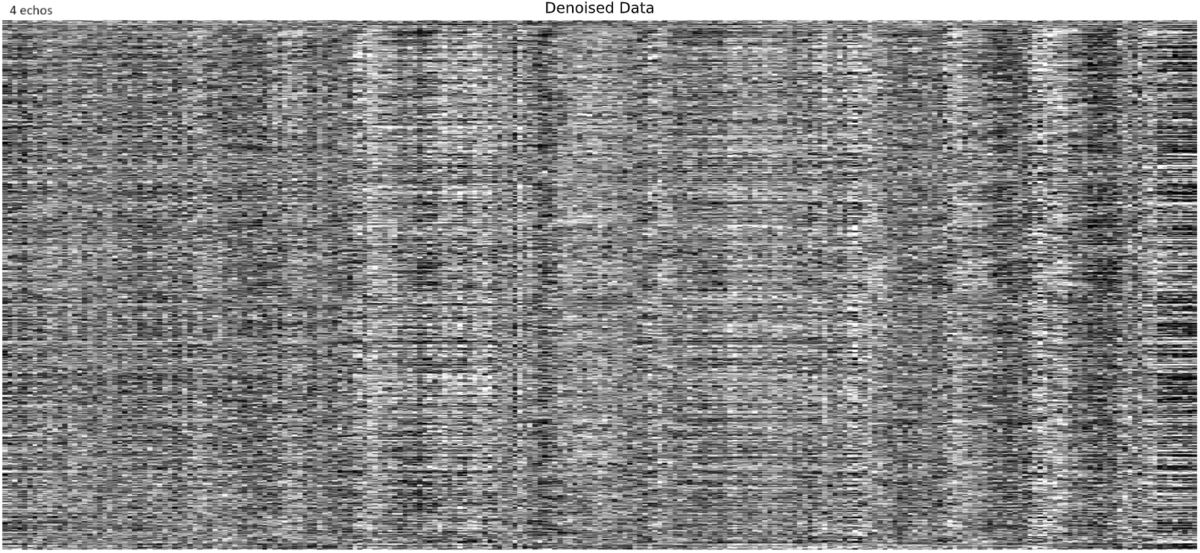
I read that completely backwards. That is a bit goofy and it is hard to pin down any specific behavior - but I agree, noise. I will say that if the noise sources like that are inaccurately retained, then polynomials in the model (or detrending/filtering) would remove the bulk of that variance - but the benefit of higher BOLD contrast due to the 4th echo being retained would remain.
These sorts of issues are also a reason tedana devs are working on some details that would allow enable more information to be passed into the model. For example, one of those artifact looks like motion to me (perhaps), and if the component had a high correlation with motion parameters, it could then be rejected in a more principled way. Alas, nothing ready quite yet.
I still think that a MB4/GRAPPA 2 4~5 echo version of your data would be closer to ideal, but I need to look over the analyses you’ve done thus far to little closer to make sure I"m not saying something too early in the morning again.
For example, one of those artifact looks like motion to me (perhaps), and if the component had a high correlation with motion parameters, it could then be rejected in a more principled way. Alas, nothing ready quite yet.
Thanks, @dowdlelt. I did see a post from @handwerkerd regarding the potential integration of both approaches.
I will look into adding another echo or two to our data (hopefully without GRAPPA, as I’m worried about subject motion during the reference scan, but we’ll see about that).
I’m less worried about motion with FLEET - its quick and somewhat robust against motion - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4580494/ but of course would still be worse than no GRAPPA in a motion case (probably - you are already using multiband which also requires a slow reference scan - the SBRef)
GRAPPA=2 would mean nearly 2x as many echoes, which is a lot of signal to leave out, imo. But I’m used to pretty low motion subjects so maybe my advice doesn’t apply here.
Thanks @dowdlelt!
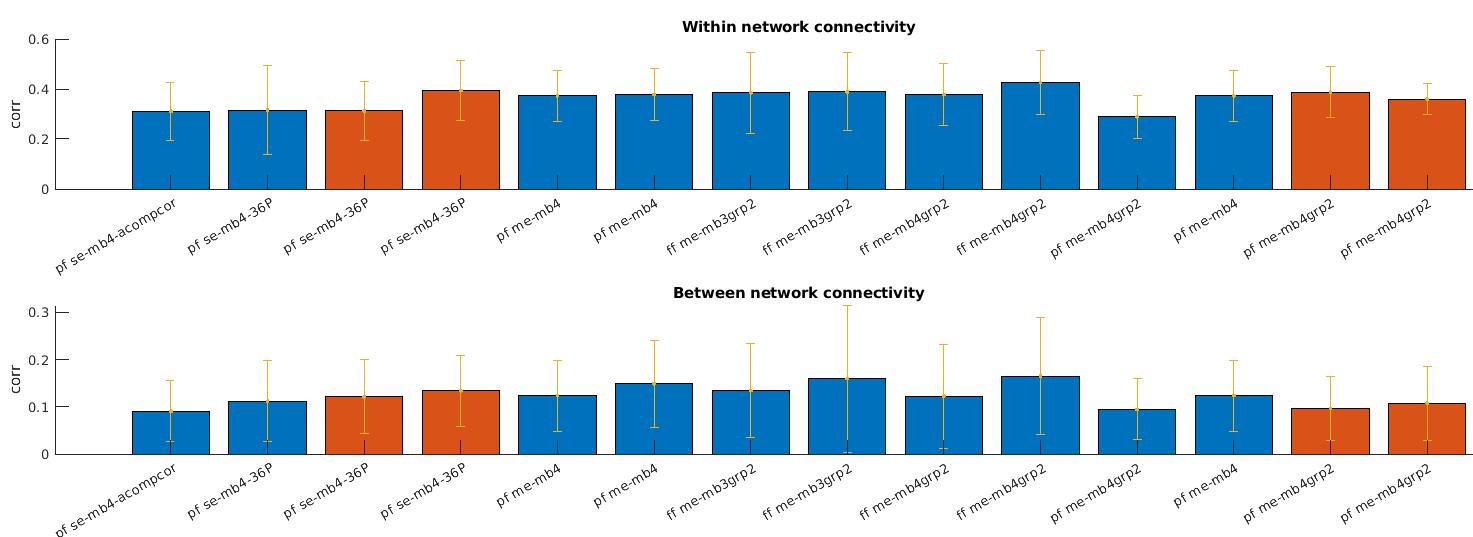
- To compare the different protocols I calculated the within-network connectivity (should be high) and between-network connectivity (presumably, should be low) for each protocol, after running tedana+xcpd with aCompCor regressors. For the single-echo scans I only applied xcpd. The two colore are two different subjects, and the single-echo scans are the first 4 bars.
se - single echo
me - multi-echo
mb - multi-band
grp - grappa
pf - partial Fourier
ff - full Fourier
Unfortunately, based only on this sample, it doesn’t seem like the mean+std of these measures improves much. Perhaps for the orange subject it does improve slightly between single-echo and multi-echo scans. Would you expect greater effects? Or is it too hard to say based on so few scans?
- I didn’t think about the time SBRef takes. You meant that noise might still leak into the data via motion during the SBRef scan, correct?
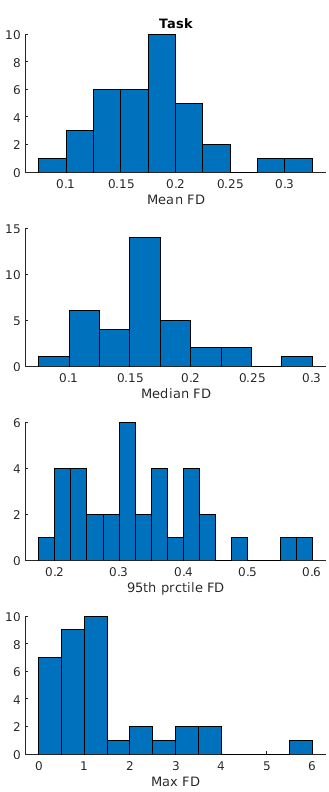
I hope this is not off-topic, but I don’t know what you consider to be low-motion for subjects. Here are some histograms shwing subject counts of several summary statistic of Framewise Displacement: in our data:
While the maxiam DF can get quite large, the median is always below 0.3mm.
Would you consider these to be low-motion?
Thanks for sticking here with me ![]()
Summary measures are nice, but as they say, garbage in, garbage out - So it could be that tedana denoising didn’t work well, or that your echoes were still too short. Things like number of echoes, last echo time, etc could all influence quality of tedana processing and results. I also think its too little data to be conclusive - but it is enough data for me to think that multiecho isn’t doing harm, which is important. Even if tedana dneoising performance isn’t perfect, that can also be adjusted for better performance - but you’ve got to have the multiecho data to start with!
Honestly though, I think this is kind of pursuing the wrong kind of final details, and its tricky to compare these things. For example: Anything with partial fourier is going to be slightly smoother, likely leading to higher correlations versus a FF scan. The full fourier scan will also have more data in hard to image regions, so potential more (but slightly noisier data) could be included.
GRAPPA is going to change distortion as well (its lower with it on), so if distortion correction isn’t applied there could be other issues with comparing.
In general, I think that 1) more echoes, and 2) echo times out to ~65ms are more important than trying to avoid GRAPPA 2, especially when using something like FLEET that is robust to that motion.
My advice, which you absolutely don’t have to follow: I think you want GRAPPA 2, and then 4 or 5 echoes out to 60ms or so. I think you can still hit your 1s target with full fourier. I know that previous data was a little odd with the noise component being left in the data, but that sort of thing is obvious to remove, and tools exists to make it easy (RICA for tedana outputs).
I just say you that you had mentioned in an earlier post that the adaptive mask had signal for all echoes - that is good - but it also means that your echo times are kind of short - even the ventral temporal lobe didn’t have enough time to lose signal! Could also explain why curve fit had different behavior - shorter echo times don’t show the full decay curve. Just a theory though.
That motion in general looks relatively low to me - all motion is bad, but that doesn’t frighten me. And yes - the worst time for someone to move is during the initial prep time of the scanner - thats when its getting the calibration scans for GRAPPA and then the SBRef and then it gets a few scans that are usually automatically discarded before starting data collection. Movement during those first two is going to be worse than movement during the scan. I think the best you can hope for is to remind participants to do their absolute best to stay still especially at the beginngin, and warn them right before scanner noises start so it doesn’t startle them.
Thanks, @dowdlelt, your responses are always so insightful!
I’m collecting a pair of short AP-PA scans prior to each run for distortion correction using top-up (I’m collecting ~10 secons of each phase encoding direction, but perhaps that’s too much and I could reduce this and use the SBRef of each phase encoding direction separately?).
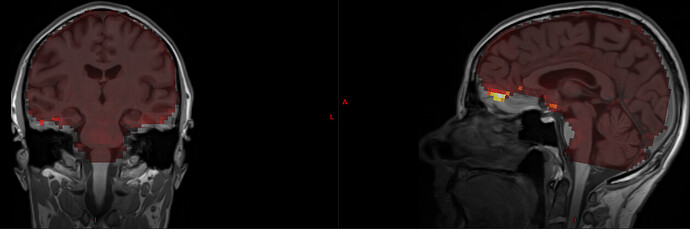
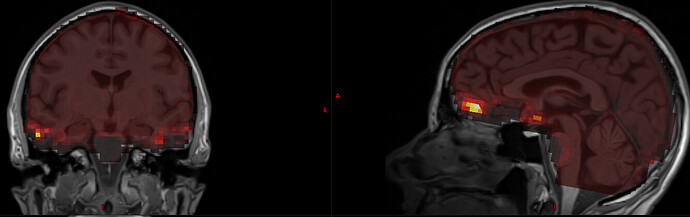
I actually already collected some data with GRAPPA2 and 4-5 echos (below are results for a protocol with 7/8 partial Fourier, MB 4, GRAPPA 2, TR-1090 ms, 3.2 mm voxels and TE=13, 27.6, 42.2, 56.8, 71.4 ms). I was surprised that the adaptive mask still had signal for all echos, even for TE=71.4 ms. I think that the issue is that the mask itself is given within the initial EPI brainmask, which excludes regions of low signal like OFC and some temporal regions. See for example here, where tedana’s S0map is overlaid on top of the T1w image:
You can also see that S0 is badly estimated at the OFC (as evident by the very high esimation values there).
I therefore reran tedana, but this time I provided it with a mask which is the union of fmriprep’s EPI brain mask and the anatomical brain mask (after regridding to EPI space). This obviously resulted in greater coverage in the S0 map:
I’m not surprised by the low S0 values there, given how low the signal is even for the first echo (13 ms):
But what surprises me is the adaptive signal mask is still perfect, and equal to 5 throughout the brain (except for a singla posterior voxel equal to 4). I don’t know what I’m missing, but that doesn’t sound quite right.
I chose an arbitrary voxel in the OFC, where the signal seems very low, and plotted the signal against the TE. Given how the linearity breaks in the log(signal) plot it seems plausible that the 5th echo would not be considered as good signal here. But perhaps the conservative way tedana calculated the adaptive signal mask still considers this “good”. Finally, there’s always the possibility that this is a voxel with non mono-exponential decay, especially given the higher probability of partial volume effects with 3.2 mm voxels.
Given that the adaptive signal mask considers these voxels “good”, would you say that combining the EPI mask with the anatomical one is a wise step, or is it too risky in terms of contaminating the data?
Thanks again for your invaluable input!
I wouldn’t worry too much about contamination - my intuition is that a mask having 5~10% more voxels (if that!) isn’t going to really break anything. I prefer to provide a liberal mask, and let tedana cut things down as it needs to.
I’m surprised about the adaptive mask bit - I’d have to peak at the data, and even then I wouldn’t be sure. I definitely wouldn’t stress over it, but it might be something we need to take a look at. If the values are so low, then they shouldn’t have a huge negative impact on the fit, but fitting to noise would be kind of annoying.
Regarding those S0 values - you’ve hit on one fact - monoexponential isn’t perfect - but even more than that, its just hard to fit the model in those regions, they are “extreme” areas. The good news is again, nothing has to be perfect for things to work well. Given that this is EPI imaging, its doesn’t need to be perfectly quantitative, just a good enough approximation for subsequent steps to work well enough.