Summary of what happened:
I import an fmriprep data set into CONN toolbox using the import feature. Then using custom python scripts I extract the top 5 anatomical component corrections from the fmriprep output, and save all of the files as .tsv files, making sure that there are no NAN or Inf values, and that the files are all the appropriate length. I then manually import all .tsv files into conn for each subject and each scan (5 scans, 2 resting state, 3 task-based). This is all in the set up tab. Then I click done, and run all subjects and all scans. When I get to the denoising tab, I get the error message that conn ran into an unexpected error (error at the end of the post).
Python code used to extract acompcor from confounds files
for sub_dir in subject_dirs:
sub = os.path.basename(sub_dir)
func_dir = os.path.join(sub_dir, "func")
if not os.path.isdir(func_dir):
print(f"No funcdir for {sub}")
continue
for file in os.listdir(func_dir):
if not file.endswith('desc-confounds timeseries.tsv'):
continue
confounds_file = os.path.join(func_dir, file)
# Load confounds
df = pd.read_csv(confounds file, sep='\t')
# Find first 5 acompcor columns
acompcor_cols = [col for col in df.columns if 'a comp_cor in col] acompcor_cols_sorted = sorted (acompcor_cols) [:5]
if not all (col in df.columns for col in acompcor_cols sorted):
print (f"Missing acompcor cols in (confounds file}")
continue
# Subset, replace Os with NaN, and convert to numeric
df subset=df[acompcor cols sorted].replace(0, pd.NA)
df_subset = df_subset.apply(pd.to_numeric, errors='coerce')
# Interpolate to fill NaNs and preserve full time series
df_clean = df_subset.interpolate (limit_direction="both").fillna(method='bfill").fillna(method='ffill')
# Save cleaned acompcor5
out_dir = os.path.join(path_output, sub)
os.makedirs (out_dir, exist_ok=True)
out_file = os.path.join(out dir, file.replace('desc-confounds_timeseries', 'acompcor5')) df clean.to_csv(out_file, sep='\t', index=False)
print (f"Saved cleaned: (out file}")
Version:
CONN version 22.2407
SPM12 version 25.01.02
Conn version from the error output says 21a because the project was started on 21a, then I downloaded and started using the newest version.
Environment (Docker, Singularity / Apptainer, custom installation):
Matlab, conn, and spm are being run in a custom virtual machine built in a type 1 hypervisor framework on a baremetal server. The VM is a linux based operating system called Red Hat Enterprise Linux, version 9.6 (Plow)
ID=“rhel”
ID_LIKE=“fedora”
Relevant log outputs (up to 20 lines):
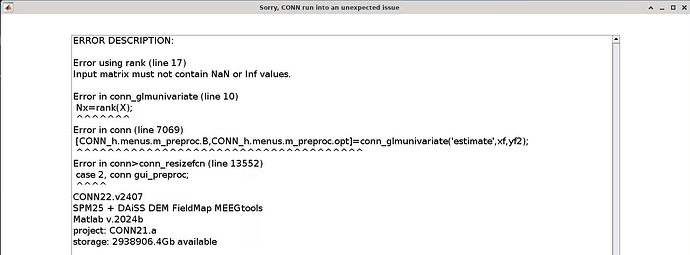
Sorry, CONN run into an unexpected issue
ERROR DESCRIPTION:
Error using rank (line 17)
Input matrix must not contain NaN or Inf values.
Error in conn_glmunivariate (line 10)
Nx rank(X);
^^^
Error in conn (line 7069)
[CONN_h.menus.m_preproc. B,CONN_h.menus.m_preproc.opt]=conn_glmunivariate('estimate', xf, yf2);
Error in conn>conn_resizefcn (line 13552)
case 2, conn gui_preproc;
CONN22.v2407
SPM25+ DAISS DEM Field Map MEEGtools Matlab v.2024b
project: CONN21.a
storage: 2938906.4Gb available