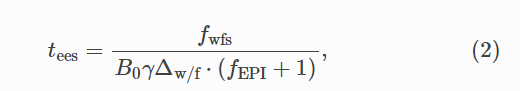Dear Experts
I am running BIDSify on some philips acquired PAR/REC images (can’t get dicoms at this point - all subjects are scanned), I had several related questions I was hoping I could get some feedack on.
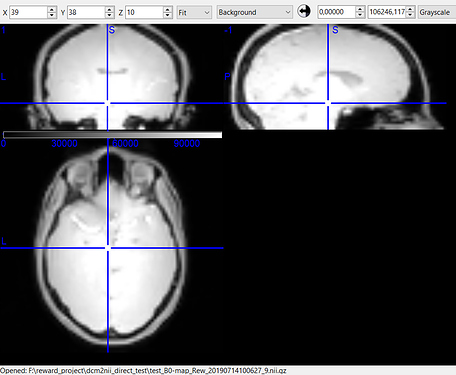
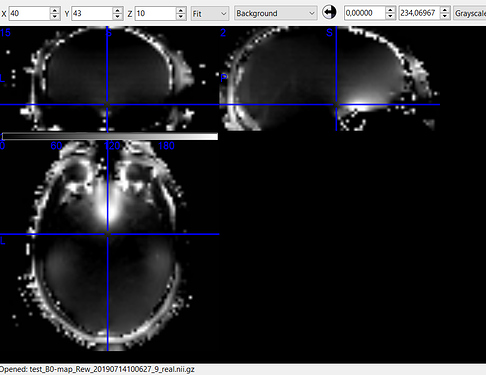
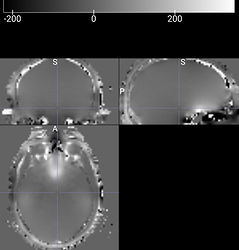
- While there was a single PAR/REC file associated with the fieldmap acquired for each bold image, it is composed of two images. When I run BIDSify, the only keys it allows to convert the file into are to a _magnitude1 and _phasediff suffix, and seems to automatically assign them. However, when I run dcm2niix directly, I get a suffixless.nii file, and a _real.nii file. Which of the two interpretations of the derived images is correct? I’ve read many forums, and @Chris_Rorden’s fieldmap page, but I’m still not sure. The first seems like either a real or imaginary file, while the second seems like either a phasediff or actual fieldmap. Here are the images. Any suggestions?
Image A - filename_magnitude1.nii (BIDSify conversion); filename.nii (direct dcm2niix conversion)
Image B - filename_phasediff.nii (BIDSify conversion); filename_real.nii (direct dcm2niix conversion)
- The next question is related to the echo times. When I run BIDSvalidator, I get the following error:
“[ERR] You have to define ‘EchoTime1’ and ‘EchoTime2’ for this file. (code: 15 - ECHO_TIME1-2_NOT_DEFINED)e[39m
./sub-005/fmap/sub-005_phasediff.nii.gz
Please visit https://neurostars.org/search?q=ECHO_TIME1-2_NOT_DEFINED”
The .json files are actually identical, except for the PhilipsRescale values. Here is the .json file for Image B.
Image B ( _real / phasediff) .json
“Manufacturer”: “Philips”,
“PatientPosition”: “HFS”,
“SeriesDescription”: “ImageMRSERIES”,
“ProtocolName”: “B0-map_Rew”,
“SeriesNumber”: 9,
“AcquisitionNumber”: 9,
“ImageComments”: “MRISCHEDELNIETSCREENEN”,
“PhilipsRescaleSlope”: 0.2442,
“PhilipsRescaleIntercept”: -500,
“PhilipsScaleSlope”: 4.095,
“UsePhilipsFloatNotDisplayScaling”: 1,
"EchoTime": 0.0032,
"RepetitionTime": 0.2,
"ReconMatrixPE": 80,
“ImageOrientationPatientDICOM”: [
0.999805,
-0.0197051,
0.00105441,
0.0197209,
0.995862,
-0.0887169 ],
“ConversionSoftware”: “dcm2niix”,
“ConversionSoftwareVersion”: “v1.0.20200331”
As mentioned, The EchoTime value of 0.0032 is the same for both images. So, assuming they ARE a magnitude and a phasediff, where would I get the Echo time 2 value, since I only have one magnitude image? Additionally, looking at the Scan Settings sheet (see box below), it says Echoes = 1. Does this mean it was not run as a multi-echo fieldmap? If so, is that a problem? What value would I put for the second echo to make this BIDS compliant?
- Relatedly, I am trying to derive the WaterFatShift EffectiveEchoSpace and TotalReadoutTime values that BIDS is asking for. I have read several posts here on neurostars, github, and jiscmail about this, and feel I am close to cracking this for my dataset. The most robust formula is possibly the following:
But I understand that there are a few settings which could make calculations unrelieable. I have values for all of the variables, but I couldn’t figure out what the ImagingFrequency variable was (I don’t have access to dicom tag numbers)… Is that the BW (Hz) field from the scanner settings sheet (i.e. 499.2)? Or a constant value (e.g. resonance frequency (MHz/T) = 42.576 from the Brainvoyager guide)
The other formula that seems to be used frequently is the OSF/Spinoza Center formula as discussed here:
where
ETL = EPI factor + 1
Here are key values from the original fieldmap .PAR file, which seems to have most of the information:
Examination name : MRI SCHEDEL NIET SCREENEN
. Protocol name : B0-map_Rew
. Examination date/time : 2019.07.14 / 10:06:27
. Series Type : Image MRSERIES
. Acquisition nr : 9
. Reconstruction nr : 1
. Scan Duration [sec] : 65.6
. Max. number of cardiac phases : 1
. Max. number of echoes : 1
. Max. number of slices/locations : 38
. Max. number of dynamics : 1
. Max. number of mixes : 1
. Patient position : Head First Supine
. Preparation direction : Anterior-Posterior
. Technique : FFE
. Scan resolution (x, y) : 80 80
. Scan mode : MS
. Repetition time [ms] : 200.000
. FOV (ap,fh,rl) [mm] : 220.000 114.675 220.000
. Water Fat shift [pixels] : 0.870
. Angulation midslice(ap,fh,rl)[degr]: 0.044 -1.132 -5.087
. Off Centre midslice(ap,fh,rl) [mm] : -5.381 30.249 -3.502
. Flow compensation <0=no 1=yes> ? : 0
. Presaturation <0=no 1=yes> ? : 0
. Phase encoding velocity [cm/sec] : 0.000000 0.000000 0.000000
. MTC <0=no 1=yes> ? : 0
. SPIR <0=no 1=yes> ? : 0
. EPI factor <0,1=no EPI> : 1
. Dynamic scan <0=no 1=yes> ? : 0
. Diffusion <0=no 1=yes> ? : 0
. Diffusion echo time [ms] : 0.0000
. Max. number of diffusion values : 1
. Max. number of gradient orients : 1
. Number of label types <0=no ASL> : 0
And here are the values from the Scanner Settings sheet, which has some more information:
ACQ voxel size RL (mm) = 2.75;
AP (mm) = 2.75;
Fold-over suppression = “no”;
Reconstruction matrix = 80;
SENSE = “no”;
k-t BLAST = “no”;
Stacks = 1;
type = “parallel”;
slices = 38;
slice gap = “default”;
slice orientation = “transverse”;
fold-over direction = “AP”;
fat shift direction = “L”;
Minimum number of packages = 1;
Slice scan order = “default”;
Patient position = “head first”;
Patient body orientation = “supine”;
Scan type = “Imaging”;
Scan mode = “MS”;
technique = “FFE”;
Fast Imaging mode = “none”;
Echoes = 1;
partial echo = “no”;
shifted echo = “no”;
TE = “shortest”;
Flip angle (deg) = 30;
TR = “user defined”;
(ms) = 200;
Halfscan = “no”;
Water-fat shift = “maximum”;
Shim = “auto”;
SAR mode = “high”;
B1 mode = “default”;
Gradient mode = “default”;
Temporal slice spacing = “default”;
fMRI echo stabilisation = “no”;
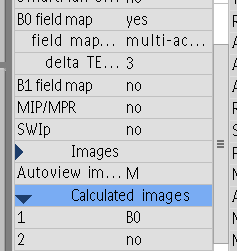
B0 field map = “yes”;
field map technique = “multi-acquisition”;
delta TE (ms) = 1;
B1 field map = “no”;
MIP/MPR = “no”;
Images = “M”, (3) “no”;
Calculated images = “B0”, (3) “no”;
Reference tissue = “Grey matter”;
Reconstruction mode = “immediate”;
Uniformity correction = “no”;
Geometry correction = “default”;
Total scan duration = “01:05.6”;
Rel. SNR = 1.05433738;
Act. TR/TE (ms) = “200 / 3.2”;
ACQ matrix M x P = “80 x 80”;
ACQ voxel MPS (mm) = “2.75 / 2.75 / 2.75”;
REC voxel MPS (mm) = “2.75 / 2.75 / 2.75”;
Scan percentage (%) = 100;
Act. WFS (pix) / BW (Hz) = “0.870 / 499.2”;
Min. WFS (pix) / Max. BW (Hz) = “0.178 / 2441.4”;
Min. TR/TE (ms) = “130 / 3.2”;
Head SAR = “< 13 %”;
Whole body SAR / level = “0.0 W/kg / normal”;
Max B1+rms = “0.83 uT”;
PNS / level = “46 % / normal”;
-
One thing that sticks out, is that the EPI_factor from the .PAR file is 1, whereas in the fucntional BOLD images, the value is 31. Is it a potential problem that EPI_factor settings do not match, when it comes time to use the fieldmap for distortion correction? Also, is EPI_factor related to SENSE? Because the second thing that jumped out was that the fieldmap acquisition was not run with SENSE, (the scan setting sheet says SENSE = “no”).
-
Finally, related to the last point, I wanted to make sure the formula I am using is correct. If no partial fourier was used, and there was no acceleration (could I be wrong about this?), would the folowing calculation based on the second formula above be valid and correct? (I did not calculate the first formula because I do not know what the imagingFrequency variable is)
effective echo spacing = (((1000 * WFS)/(434.215 * (ETL+1))/acceleration)
= (1000 * 0.87)/(434.215 * 3) = 0.668
Where
WaterFatShift = 0.87
ETL = EPI_Factor + 1 = 2
No acceleration
And
TotalReadOutTime = EffectiveEchoSpace * 79 = 52.76
Where
ReconMatrixPE = 80
Sorry for the lenght, but I think including all this information may be helpful to other Philips users struggling with this same issue. Thanks ahead for any observations/comments!!
Best,
Carlos