Hello,
I am new to fMRI data analysis and I would like to ask a question about the quality of the masks I am obtaining.
I run CPAC on a patient using the default parameters, and opening the files
space-MNI152NLin6ASym_label-GM_mask.nii.gz (see the GIF below)

and
space-MNI152NLin6ASym_label-WM_mask.nii.gz (see the GIF below)

I noticed that both of them present some parts that shouldn’t be there, like parts of the skull.
However, if for example I look at the (functional) image task-func1_space-MNI152NLin6ASym_reg-default_desc-smZstd1_reho.nii.gz
the artifact are not present and it seems there is nothing wrong with it.
Nonetheless, since we have the idea of doing a similar preprocessing to an entire dataset of images, and thus a control for each generated output is not feasible, I wanted to ask whether there is an automatic way of checking this masks in order to understand if CPAC worked well, producing masks without artifacts.
Also, is there any way to know if these kinds of artifacts can affect the analysis outputs?
I thank you in advance for your help!
Hi, thank you for reaching out!
I know you said you used default settings, but could you provide your pipeline file just so I can see? The non-skullstripped image should not be going through segmentation, so there might be an issue with the skullstripping settings.
I wanted to ask whether there is an automatic way of checking this masks in order to understand if CPAC worked well, producing masks without artifacts.
Also, is there any way to know if these kinds of artifacts can affect the analysis outputs?
We have a few QC outputs that can be configured under output_directory > quality_control > generate_quality_control_images, although these qc measures don’t provide visualizations of the tissue masks. The best way to ensure they are being created properly is to visualize them when processing completes. And yes, if segmentation isn’t working properly then this can definitely affect the outputs.
Thank you for your reply!
You find attached the pipeline file
pipeline_config.txt (69.8 KB)
Hi, sorry for the delay! Everything looks good with the skullstripping settings. Could you also post your skull-stripped brain? It will be called either desc-preproc_T1w or desc-brain_T1w. I would like to see if there are still sections of skull after masking.
Hi, no worries! Thank you for your help.
You find below the image you requested.
space-MNI152NLin6ASym-desc-preproc_T1w
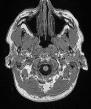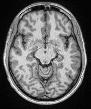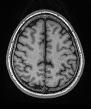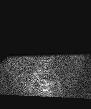
Let me know if it was what you were asking for or you meant some other image.
Interesting, it looks like skullstripping didn’t run for the T1. Are there any crash files in your /log folder?
Hi! There are no crash files. In the log folder we have these files:
Thank you! Could you post the pypeline.log file? I want to see if skullstripping actually ran.
Hello! You find attached the pypeline.log file. Thank you for your time!
pypeline.txt (1.5 MB)
It looks like skullstrip ran successfully, so there may be a data-specific issue with the default parameters for BET. I would suggest increasing the fractional intensity threshold, since the threshold may be too low for this data. The BET user guide says that a higher threshold (>.5) results in a smaller segmented brain. If the skullstripping quality is still poor with the adjusted parameters, you could try a different skullstripping tool. I am also happy to test this out for you if the dataset you’re using is publicly available!
Please let me know how this goes or if you have any questions.
Hi, thank you! Unfortunately I cannot share our dataset. I’ll try to do what you suggest and let you know how it goes
Hi! I tried changing the fractional intensity threshold to 0.6 and also to 0.75 but the skullstripping quality is still poor. Should I try an higher value or maybe a smaller one? If not, what skullstripping tool would you recommend?
Hi! You could definitely try increasing the threshold more, but I would recommend forking the different skullstripping options so that you can see which works best for your data. You can configure this in the pipeline file here. I recommend this:
using: ['3dSkullStrip', 'UNet', 'niworkflows-ants']
Since listing more than one option will result in multiple parallel forks, you can look in the json metadata to see which file used which skullstripping method.
Let me know if you have any trouble!