Hi everyone!
I’m hoping someone else has ran into this issue and knows where the problem is occurring.
I am working with abcd data, using the
sub-*_ses0**_task-rest_bold_desc-filtered_timeseries.dtseries.nii
file.
We want to remove the subcortical structures in order to run ICA + DR on purely the cortical data (CORTEX_LEFT and CORTEX_RIGHT). This is successfully achieved using the following commands:
wb_command -cifti-separate input.nii COLUMN -metric CORTEX_LEFT left.func.gii -metric CORTEX_RIGHT right.func.gii
wb_command -cifti-create-dense-scalar cortex_only.nii -left-metric left.func.gii -right-metric right.func.gii
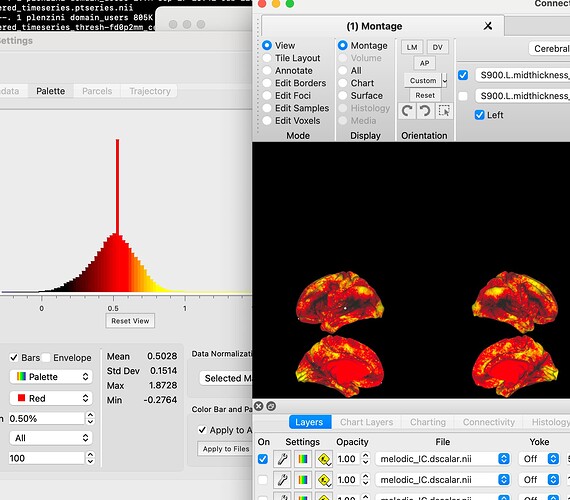
However, when Melodic (ICA) is run on the resulting files, there is uniform, non-zero data in the medial wall of the ICA components. Notably, they are not the same values between the components. I have attached a photo of one of the most striking examples. I cannot find any information in the documentation of the commands, but I thought perhaps someone here would know where the issue is occurring.
Thanks for your time in advance!
Angelica