Hi,
Entire code:
Hi,
Got it. Another question. I thought the participant.tsv was supposed to be
comprised of all the sub-* participants in the BIDS directory and it updates with each subject.
I ran the initial scaffold and it created this:
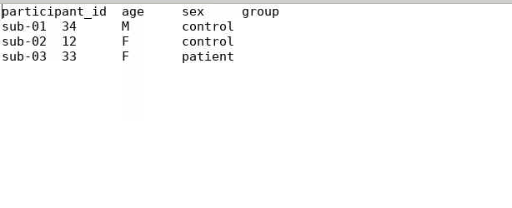
Then when I run the dcm2bids conversion it doesn’t update the participant.tsv file.
I get the nii.gz and json files for the subject that I tested but the .tsv file does not update.
**P.S. Sorry for all the questions. I much more familiar with heudiconv and less familiar with dcm2bids.
**I ran the scaffold first from one shell script and then the dcm2bids singularity command from another
shell script.
–Thanks.
Subbi M.
The scaffold script does not update the participants tsv, it has no way of knowing your subject demographics.
Hi all,
I’m running into an error for my config file conversion. The series description for the DICOM data that I am converting is not matching up and it is giving error of multiple pairings without the nifti file being generated.
My log file is:
The file that I am referring to is FMRI-NX1-STT2-PA. Its giving multiple pairing under T2w and bold but the nifti file is not converted. There is no nii.gz file for STT2-PA. My config file logic is:
T2w conversion logic in my config file is:

Note: task-mid and task-nback both have a similar logic to task-sst and they are getting converted just fine.
Anything anybody else is seeing for why this error is occuring?
Thanks.
–Subbi M.
If you could post code-formatted text instead of screenshots in the future that would be appreciated.
The issue is that your T2w criteria is not specific enough, as BOLD file SeriesDescription contains T2 in SIT2.
Best,
Steven
Hi @Steven,
Thanks–noted about code formatted text. Will revise in the future.
Thank you! Will revise and see if there are any more errors.
Thanks.
–Subbi M.
MANAGED BY INCF