oups. I fixed the command line above. Please check and make sure I put the right path to dcm2bids_dev.sif
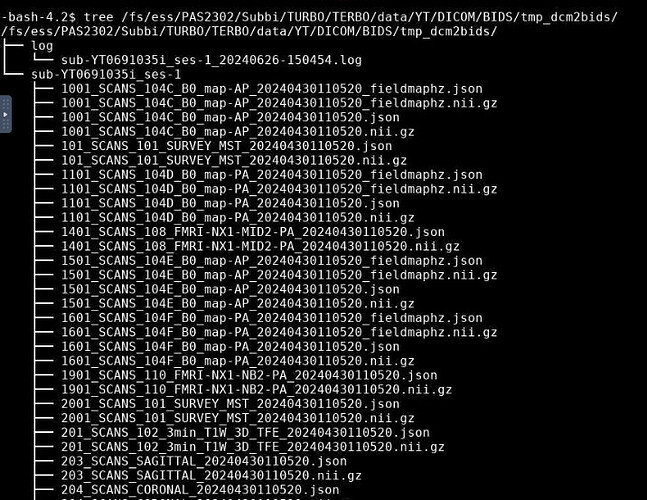
Hi @abore Okay we are making progress. The dicoms have now been converted to Nifti but still not BIDS format. Example of tree for subject
The log that the command prints is really long. But this is the last thing that prints on the CLI when the singularity command is executed.
INFO | SIDECAR PAIRING
WARNING | Several Pairing ← 101_SCANS_101_SURVEY_MST_20240430110520
Traceback (most recent call last):
File “/venv/bin/dcm2bids”, line 8, in
sys.exit(main())
^^^^^^
File “/dcm2bids/dcm2bids/cli/dcm2bids.py”, line 125, in main
app = Dcm2BidsGen(**vars(args)).run()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File “/dcm2bids/dcm2bids/dcm2bids_gen.py”, line 108, in run
parser.build_acquisitions(self.participant)
File “/dcm2bids/dcm2bids/sidecar.py”, line 416, in build_acquisitions
acq = Acquisition(participant,
^^^^^^^^^^^^^^^^^^^^^^^^
TypeError: Acquisition.init() missing 2 required positional arguments: ‘datatype’ and ‘suffix’
(dcm2bids)
Do you want me to share the entire log on here?
No need to share it I think I know the next step.
Please make sure your config.file follow the new dcm2bids convention.
Use this link.
Then try again ![]()
Hi,
So I have several folders that don’t actually need to be converted to BIDS format that is within the SCANS folder. After the SCANS folder there are multiple subfolders then within each of those subfolders there is a DICOM folder. I just need to convert the folders that have a subfolder of DICOM within SCANS. There are several warning that are printing:
-
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed.
2024-06-26 15:22:08.555 - WARNING - sidecar.isLink | This key contains is not allowed. -
2024-06-26 15:22:08.594 - WARNING - sidecar.build_acquisitions | → _bold
2024-06-26 15:22:08.594 - WARNING - sidecar.build_acquisitions | Several Pairing ← 1001_SCANS_104C_B0_map-AP_20240430110520_fieldmaphz
2024-06-26 15:22:08.594 - WARNING - sidecar.build_acquisitions | → _T1w
2024-06-26 15:22:08.594 - WARNING - sidecar.build_acquisitions | → _T2w
2024-06-26 15:22:08.595 - WARNING - sidecar.build_acquisitions | → _bold
2024-06-26 15:22:08.595 - WARNING - sidecar.build_acquisitions | → _bold
2024-06-26 15:22:08.595 - WARNING - sidecar.build_acquisitions | → _bold
2024-06-26 15:22:08.595 - WARNING - sidecar.build_acquisitions | Several Pairing ← 1101_SCANS_104D_B0_map-PA_20240430110520 -
2024-06-26 15:22:08.618 - WARNING - dcm2bids_gen.run | NO PAIRING WAS FOUND. BIDS FOLDER “/FS/ESS/PAS2302/SUBBI/TURBO/TERBO/DATA/YT/DICOM/BIDS/SUB-YT0691035I/SES-1” WON’T BE CREATED. CHECK YOUR CONFIG FILE.
2024-06-26 15:22:08.618 - INFO - dcm2bids.main | Logs saved in /fs/ess/PAS2302/Subbi/TURBO/TERBO/data/YT/DICOM/BIDS/tmp_dcm2bids/log/sub-YT0691035i_ses-1_20240626-152155.log
2024-06-26 15:22:08.618 - INFO - dcm2bids.main | — dcm2bids end —
My updated config file looks like this:
{
“descriptions”: [
{
“datatype”: “anat”,
“suffix”: “T1w”,
“criteria”: {
“SeriesDescription”: {
“contains”: “T1W”
}
}
},
{
“datatype”: “anat”,
“suffix”: “T2w”,
“criteria”: {
“SeriesDescription”: {
“contains”: “T2”
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: “task-sst_dir-PA”,
“criteria”: {
“SeriesDescription”: {
“contains”: “STT2”
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: “task-sst_dir-AP”,
“criteria”: {
“SeriesDescription”: {
“contains”: “STT1”
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: “task-mid_dir-PA”,
“criteria”: {
“SeriesDescription”: {
“contains”: “MID2”
}
}
}
]
}
This is not something dcm2bids allows:
“criteria”: {
“SeriesDescription”: {
“contains”: “MID2”}
}
If you found this somewhere in the documentation please tell so I can fix it.
Check here to find the correct way to write your config file.
The option -d should point to the folder containing the dicoms you want to convert. You can also be more specific with your criteria in the config file, it will convert your dicom into nifti files but it will reorder only the files you need.
Hi @abore Okay. Thanks. I’ll update my config file, and re-run it latest by tomorrow. I’ll let you know if there are any issues.
Thanks.
–Subbi M.
Hi,
Separate question. If I want the folder structure to be sub-{subject}/ses-{session} instead of sub-{subject}_ses-{session} is that a change in the Shell script or the config file?
Thanks.
–Subbi M.
The output structure created by dcm2bids is: sub-{subject}/ses-{session}/
Once you fixed your config file it should work as expected.
Hi @abore
How do I make this more specific:
{
"dataType": "func",
"modalityLabel": "bold",
"customLabels": "task-sst",
"criteria": {
"SeriesDescription": "*STT1*"
}
},
Let’s say I have 2 runs of task-sst. Ignore that the seriesdescription says STT-they misspelled it in the scanner. STT1 is run-01 and STT2 is run-02.
I want my end .nii.gz file and .json file to look like this:
sub-YT7060217L_ses-1_task-sst_dir-AP_run-01_bold.nii.gz
Also where do I put acq-label if I want to customize the above further?
Please advice on how to edit the config file so my BIDS data looks like the naming convention above.
Thanks.
*Also I know I’m working of an older version will revise it to fit newer config.json file standard labels at the end. Thanks.
–Subbi M.
Hello,
You can hardcode the run and its number in the custom entities like this “run-01” for this description and add another one with STT2.
You can add more custom entities. Check this link.
Arnaud
Hi,
Is there a way to modify the config file so it doesn’t hard code the runs. So for example in heudiconv doing the heuristic:
mid = create_key(‘sub-{subject}/{session}/func/sub-{subject}_{session}_task-mid_run-{item:02d}_bold’)
The run-{item:02d}_bold takes the first occurrence of a task and labels it as run-01 and the second occurrence of a task and labels it as run-02 automatically.
Is there a way to do that with the newest version of dcm2bids so I don’t have to hard-code it. I don’t know for certain how many runs there are of a task. Sometimes in a scanner if run-01 is partial they will sometimes re-do it.
I don’t want to hardcode it.
Thanks.
–Subbi M.
Hello,
It’s exactly how dcm2bids works but you don’t have to do anything.
Change your config file like this:
"SeriesDescription": "*STT*"
Then the first iteration will be named run-01 and the second one run-02.
Arnaud
Hi,
When I run the singularity command it generates the nii.gz and json sidecar files but its not generating the BIDS tree structure.
scaffold_directory/
├── CHANGES
├── code/
├── dataset_description.json
├── derivatives/
├── participants.json
├── participants.tsv
├── README
├── .bidsignore
└── sourcedata/
How do I generate the participants.tsv and other json files. It says dcm2bids generates this automatically but its not generating the files I need to run MRIQC. Do I need to add something to my config.json?
Thanks.
--Subbi M.Without seeing your most recent config, command, and log, there’s not much we can do to help.
Best,
Steven
Hi,
Here is my config.json
{
“descriptions”: [
{
“datatype”: “anat”,
“suffix”: “T1w”,
“custom_entities”: “run-01”,
“criteria”: {
“SeriesDescription”: “T1W_3D_TFE”
}
},
{
“datatype”: “anat”,
“suffix”: “T2w”,
“custom_entities”: “run-01”,
“criteria”: {
“SeriesDescription”: “T2”
}
},
{
“datatype”: “fmap”,
“suffix”: “epi”,
“custom_entities”: “dir-AP”,
“criteria”: {
“SeriesDescription”: “*B0 map-AP”
}
},
{
“datatype”: “fmap”,
“suffix”: “epi”,
“custom_entities”: “dir-PA”,
“criteria”: {
“SeriesDescription”: “*B0 map-PA”
}
},
{
“datatype”: “fmap”,
“suffix”: “fieldmap”,
“custom_entities”: “acq-B0map”,
“criteria”: {
“SeriesDescription”: “*B0 map”,
“ImageType”: [“ORIGINAL”, “PRIMARY”, “T1”, “MIXED”, “REAL”, “FIELDMAPHZ”]
}
},
{
“datatype”: “fmap”,
“suffix”: “magnitude”,
“custom_entities”: “acq-B0map”,
“criteria”: {
“SeriesDescription”: “*B0 map”,
“ImageType”: [“ORIGINAL”, “PRIMARY”, “T1”, “MIXED”, “FIELDMAPHZ”]
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-gamb”, “dir-AP”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRIgamb-AP”, “*1NX_fMRI_gamb-AP”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-gamb”, “dir-PA”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRIgamb-PA”, “*1NX_fMRI_gamb-PA”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-wm”, “dir-AP”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRI_wm-AP”, “*1NX_fMRI_wm-AP”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-wm”, “dir-PA”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRI_wm-PA”, “*1NX_fMRI_wm-PA”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-rest”, “dir-AP”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRI_rs-AP”, “*1NX_fMRI_rest-AP”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-rest”, “dir-PA”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRI_rs-PA”, “*1NX_fMRI_rest-PA”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-ep”, “dir-AP”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRI_ep-AP”, “*1NX_fMRI_ep-AP”]
}
}
},
{
“datatype”: “func”,
“suffix”: “bold”,
“custom_entities”: [“task-ep”, “dir-PA”],
“criteria”: {
“SeriesDescription”: {
“any”: [“*HCP_fMRI_ep-PA”, “*1NX_fMRI_ep-PA”]
}
}
},
{
“datatype”: “dwi”,
“suffix”: “dwi”,
“custom_entities”: [“acq-dti”, “dir-PA”],
“criteria”: {
“SeriesDescription”: “MB4_102d_PA”
}
},
{
“datatype”: “dwi”,
“suffix”: “dwi”,
“custom_entities”: [“acq-B0”, “dir-PA”],
“criteria”: {
“SeriesDescription”: “DTI B0 PA”,
“ImageType”: [“ORIGINAL”, “PRIMARY”, “DIFFUSION”, “NONE”]
}
},
{
“datatype”: “dwi”,
“suffix”: “dwi”,
“custom_entities”: [“acq-B0”, “dir-AP”],
“criteria”: {
“SeriesDescription”: “DTI B0 AP”,
“ImageType”: [“ORIGINAL”, “PRIMARY”, “DIFFUSION”, “NONE”]
}
}
]
}
Script I’m running:
#!/bin/bash
#SBATCH --time=01:00:00
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=2
#SBATCH --account=PAS2302
folder=$1
subject=$2
session=$3
echo “Starting BIDS conversion for subject” $folder $subject $session
singularity exec
/fs/ess/PAS2302/Subbi/TURBO/TERBO/Scripts/dcm2bids_dev.sif
dcm2bids
-d /fs/ess/PAS2302/Subbi/TURBO/TERBO/data/YA/DICOM/$folder/SCANS/*
-p $subject
-s $session
-o /fs/ess/PAS2302/Subbi/TURBO/TERBO/data/YA/DICOM/BIDS
-c /fs/ess/PAS2302/Subbi/TURBO/TERBO/Scripts/study_config_YA.json
–clobber
echo “Finished subject” $subject $session
This is the log:
2024-07-03 15:01:29.679 - INFO - dcm2niix_gen.execute | Check log file for dcm2niix output
2024-07-03 15:01:29.721 - INFO - sidecar.build_acquisitions | SIDECAR PAIRING
2024-07-03 15:01:29.721 - INFO - sidecar.build_acquisitions | No Pairing ← 101_101_199_SURVEY_MST_199_SURVEY_MST_20231121100251
2024-07-03 15:01:29.721 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_run-01_T1w ← 201_201_198_3min_T1W_3D_TFE_198_3min_T1W_3D_TFE_20231121100251
2024-07-03 15:01:29.721 - INFO - sidecar.build_acquisitions | No Pairing ← 203_203_SAGITTAL_SAGITTAL_20231121100251
2024-07-03 15:01:29.721 - INFO - sidecar.build_acquisitions | No Pairing ← 204_204_CORONAL_CORONAL_20231121100251
2024-07-03 15:01:29.722 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_dir-AP_epi ← 301_301_197_B0_map-AP_197_B0_map-AP_20231121100251
2024-07-03 15:01:29.722 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_dir-AP_epi ← 301_301_197_B0_map-AP_197_B0_map-AP_20231121100251_fieldmaphz
2024-07-03 15:01:29.722 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_dir-PA_epi ← 401_401_101_B0_map-PA_101_B0_map-PA_20231121100251
2024-07-03 15:01:29.722 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_dir-PA_epi ← 401_401_101_B0_map-PA_101_B0_map-PA_20231121100251_fieldmaphz
2024-07-03 15:01:29.722 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_task-ep_dir-PA_bold ← 601_601_104_HCP_fMRI_ep-PA_104_HCP_fMRI_ep-PA_20231121100251
2024-07-03 15:01:29.722 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_task-wm_dir-AP_bold ← 801_801_105B_1NX_fMRI_wm-AP_105B_1NX_fMRI_wm-AP_20231121100251
2024-07-03 15:01:29.724 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_task-wm_dir-PA_bold ← 1201_1201_102B_1NX_fMRI_wm-PA_102B_1NX_fMRI_wm-PA_20231121100251
2024-07-03 15:01:29.724 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_task-rest_dir-AP_bold ← 1301_1301_106B0_HCP_fMRI_rs-AP_106B0_HCP_fMRI_rs-AP_20231121100251
2024-07-03 15:01:29.724 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_task-rest_dir-AP_bold ← 1601_1601_106B0_HCP_fMRI_rs-AP_106B0_HCP_fMRI_rs-AP_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_acq-B0map_magnitude ← 1701_1701_107_B0_map_107_B0_map_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_acq-B0map_fieldmap ← 1701_1701_107_B0_map_107_B0_map_20231121100251_fieldmaphz
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_run-01_T2w ← 1801_1801_111_3D_T2_SAG_RAW_111_3D_T2_SAG_RAW_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | No Pairing ← 1803_1803_MPR_COR_0.8_-0.4mm_MPR_COR_0.8_-0.4mm_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | No Pairing ← 1804_1804_MPR_AX_0.8_-0.4mm_MPR_AX_0.8_-0.4mm_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_acq-B0_dir-PA_dwi ← 1901_1901_114_DTI_B0_PA_114_DTI_B0_PA_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | No Pairing ← 1903_1903_DIFF_SAG_DIFF_SAG_20231121100251
2024-07-03 15:01:29.725 - INFO - sidecar.build_acquisitions | No Pairing ← 1904_1904_DIFF_COR_DIFF_COR_20231121100251
2024-07-03 15:01:29.726 - INFO - sidecar.build_acquisitions | sub-YA0690246B_ses-1_acq-B0_dir-AP_dwi ← 2001_2001_114_DTI_B0_AP_114_DTI_B0_AP_20231121100251
2024-07-03 15:01:29.726 - INFO - sidecar.build_acquisitions | No Pairing ← 2003_2003_DIFF_SAG_DIFF_SAG_20231121100251
2024-07-03 15:01:29.726 - INFO - sidecar.build_acquisitions | No Pairing ← 2004_2004_DIFF_COR_DIFF_COR_20231121100251
2024-07-03 15:01:29.726 - INFO - sidecar.find_runs | sub-YA0690246B/ses-1/fmap/sub-YA0690246B_ses-1_dir-AP_epi has 2 runs
2024-07-03 15:01:29.728 - INFO - sidecar.find_runs | Adding run information to the acquisition
2024-07-03 15:01:29.728 - INFO - sidecar.find_runs | sub-YA0690246B/ses-1/fmap/sub-YA0690246B_ses-1_dir-PA_epi has 2 runs
2024-07-03 15:01:29.728 - INFO - sidecar.find_runs | Adding run information to the acquisition
2024-07-03 15:01:29.729 - INFO - sidecar.find_runs | sub-YA0690246B/ses-1/func/sub-YA0690246B_ses-1_task-rest_dir-AP_bold has 2 runs
2024-07-03 15:01:29.729 - INFO - sidecar.find_runs | Adding run information to the acquisition
2024-07-03 15:01:29.729 - INFO - dcm2bids_gen.run | Moving acquisitions into BIDS folder “/fs/ess/PAS2302/Subbi/TURBO/TERBO/data/YA/DICOM/BIDS/sub-YA0690246B/ses-1”.
2024-07-03 15:01:29.784 - INFO - dcm2bids.main | Logs saved in /fs/ess/PAS2302/Subbi/TURBO/TERBO/data/YA/DICOM/BIDS/tmp_dcm2bids/log/sub-YA0690246B_ses-1_20240703-150111.log
2024-07-03 15:01:29.784 - INFO - dcm2bids.main | — dcm2bids end —
Apologies I read your post too fast. You need the dcm2bids_scaffold command to generate all of the BIDS files and folders. But in future if your are going to be pasting lots of text from the command line please use tick marks so the text is nicely formatted like this or
Like this
Best,
Steven
Hi,
If I’m using a singularity dcm2bids sif command do I add the scaffold before it?
singularity exec
/fs/ess/PAS2302/Subbi/TURBO/TERBO/Scripts/dcm2bids_dev.sif
dcm2bids \
Add dcm2bids_scaffold before dcm2bids or does it need to be done separately beforehand?
Also you text suggestion formatting is not coming up. Do I click the blockquote to reformat my text when replying?
–Subbi M.
singularity exec \
-e --containall -B $bids \
/fs/ess/PAS2302/Subbi/TURBO/TERBO/Scripts/dcm2bids_dev.sif \
dcm2bids_scaffold $bids
It’s the </> button in the text editor (or use the ` button on your keyboard)
Best,
Steven
Hi,
I ran it for the initial subject and it worked but then when I tried running dcm2bids for a second subject
I got an error for scaffolding. Do I have to use the force button?
Is this right?
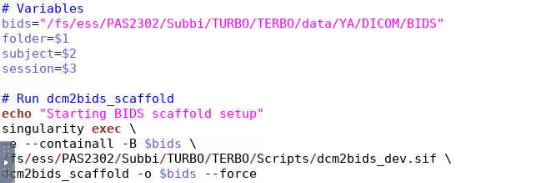
Command in command line: ./osc_convert2nifti_TERBO_YA_Bronx.sh $foldername $subject_id $session_id.
Thanks.
--Subbi M.