I have been obtaining nice results when defacing (T1) MRI images using mri_deface and pydeface approaches. Nevertheless, both algorithms return a .nii image, which is not always desirable (sometimes one need to share DICOM files instead of .nii with collaborators).
I am trying to develop a pipeline to deface DICOM images. The idea is to go from DICOM to Nifti (dcm2niix), deface (mri_deface) and then generate synthetic DICOMs using the original DICOM file and modifying the PixelData file.
The problem I am facing is two-fold:
- Which DICOM field might give me information on the direction/orientation the 2-D slices were acquired? (i.e it was sagittal? Axial?)
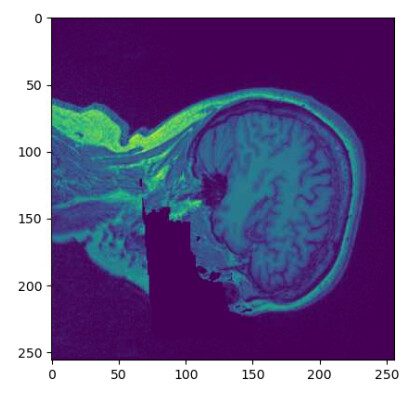
- I am struggling with orientation… Whem I load the defaced nifti file using nibabel (left), I can not get the same orientation than the data stored on PixelData (right).
Image I obtain when plotting the dicom PixelData (pydicom.dcmread(infile).pixel_array):
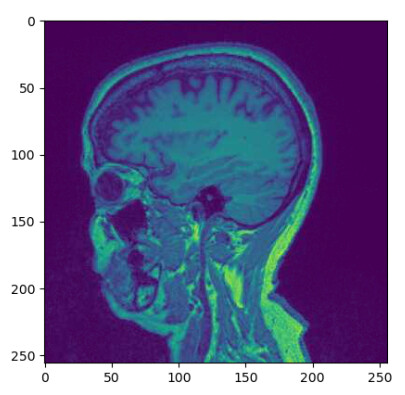
Image I obtain when plotting the defaced nifti image:
Any insight? Does this re-orientation of the defaced image has an impact AT ALL when storing the bytes on the DICOM file?
Thanks!
@slieped are you able to control the DICOM-to-NIfTI conversion, or must you deal with data that is already in NIfTI format?
If you are able to control DICOM-to-NIfTI conversion, you may want to try to convert your files with the -x i and -y n option, e.g. dcm2niix -x i -y n /path/to/DICOMs. By default, dcm2niix will losslessly rotate 3D acquisitions to be RAS aligned. The -x i ignores this transform. For DICOMs, the first line of the image is at the top of the screen, with subsequent lines beneath, the way we read English. In contrast, with NIfTI the first line is at the bottom of the screen, with larger values above, the way we draw the y axis of a cartesian graph. Therefore, by default dcm2niix reverses the order of lines. The -y n disables the reversal of the y coordinate. For more details see the dcm2niix notes on spatial coordinates.
If you are dealing with existing NIfTI files, you will want to examine the dcm2niix notes on spatial coordinates to understand the intrinsic differences between these two formats. The NIfTI format does not have all the sequence data required for a legal DICOM modality MR, so you will want to make your DICOMs of modality other (OT). You will also have to generate UIDs and transform the NIfTI spatial coordinates to the DICOM method. A quick web search shows several tools that can help in this process.
1 Like
Thank you very much @Chris_Rorden !!
I can actually control the DCM 2 nifti. I will play around with the -x i option, but it sounds exactly what I need.
Again, thanks for your excellent work with dcm2niix and, to always answer with detailed responses.

