Good evening fMRI Experts,
I having been working on a PPI analysis with Human Connectome Project for Early Psychosis (HCP-EP) resting state data. In making the confound files for the first level analysis (which includes denoising) I noticed some subjects’ confound files were of varying sizes based on the presence (or lack thereof) of non-steady state outlier columns. Most subjects/runs do not have non-steady state confounds while some do. I am under the impression this is based on the initial detection of outliers early in the scan (not necessarily whether or not they are actually dummy scans), and that their identification may signal something wrong with the data.
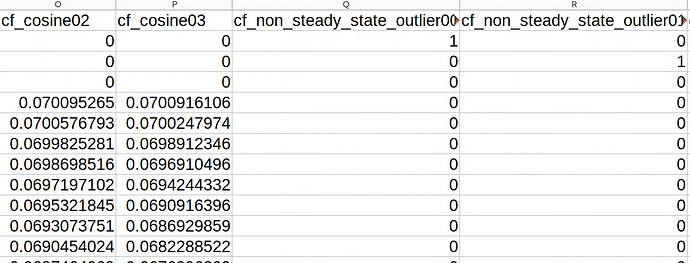
I found this NeuroStars post on the subject, but I am not sure if I am understanding correctly. It seems that when these outliers are detected, the algorithm terminates the estimation of CompCors, and these rows are just 0’s. Here is what it looks like:
My research has suggested that the HCP consortium discards non-steady state volumes in their distributions. When we ran fMRIprep, we did not include the --dummy-scans 0 argument mentioned in the previous NeuroStars post I linked, and because of this I am wondering which of the following courses of action would be the most appropriate:
-
Rerun fMRIprep with the --dummy-scans 0 argument, and redo the whole analysis
-
Keep things as they are and “regress out” the non_steady_state outlier columns in the first level analysis. I am wondering if this would be statistically reasonable since each non-steady state outlier column lists a single 1 flagging the offending volume and is padded everywhere else with 0’s.
-
Should I simply exclude runs for which there are any non_steady_state outliers for in first level analysis?
I sincerely appreciate any insights the community may be able to offer.
Kind regards,
Linda