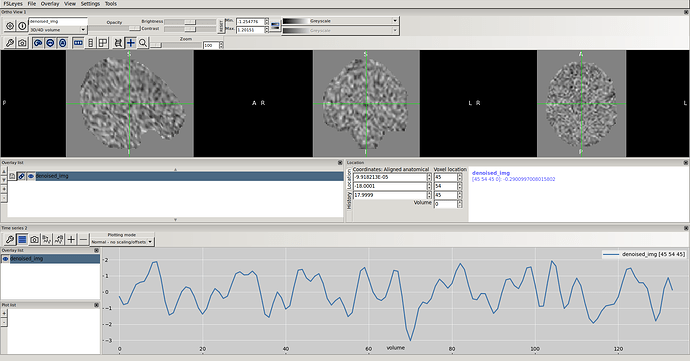
Hi everyone,
Now I am trying to denoise my data and conduct first level analysis on this denoised data. I am using nipype workflow and nodes, SPM first level analysis and nilearn clean_img function. In addition, I am using physiological regressors calculated by PhysIO toolbox. So here is my code
def denoise_img(func_path, mask_path, phys_regressors, bids_dir):
import nibabel as nib
import pandas as pd
from nilearn import image as niimg
import os
denoise_path = os.path.join(bids_dir, "derivatives", "fMRI_Denoise", "denoised_img.nii")
phys_regressors = phys_regressors.to_numpy()
clean_img = niimg.clean_img(func_path,confounds= phys_regressors, axis=0,detrend=True,standardize=True,
low_pass=0.1,high_pass=0.009,t_r= 2, mask_img=mask_path)
nib.save(clean_img, denoise_path)
return denoise_path
def physio_toolbox(script_mat, subject, session, task, table, derivatives_dir):
from nipype.interfaces.matlab import MatlabCommand
import pandas as pd
filtered_data = table[(table['subject'] == subject) & (
table['session'] == session) & (table['task'] == task)]
tsv_paths = None
tsv_path = ''
if not filtered_data.empty:
tsv_paths = filtered_data.iloc[0]['tsv_physio']
# Check if the list is not empty before accessing the first item
if tsv_paths:
tsv_path = tsv_paths[0]
with open(script_mat, 'r') as file:
script_mat = file.read()
script_mat = script_mat.replace('physio.log_files.cardiac = {};', f"physio.log_files.cardiac = {{'{tsv_path}'}};").replace("physio_out",
f"{derivatives_dir}/sub-{subject}/ses-{session}/physio_out_{task}")
if task == "selfref":
script_mat = script_mat.replace("Nscans = 575", "Nscans = 545")
elif task == "faces":
script_mat = script_mat.replace("Nscans = 575", "Nscans = 134").replace("TR = 0.735", "TR = 2").replace("Nslices = 64", "Nslices = 34").replace("onset_slice = 32", "onset_slice =17")
elif task == "reversal":
script_mat.replace("Nscans = 575", "Nscans = 1820")
matlab = MatlabCommand()
matlab.inputs.script = script_mat
matlab.run()
phys_regressors = f'{derivatives_dir}/sub-{subject}/ses-{session}/physio_out_{task}/multiple_regressors.txt'
with open(phys_regressors, 'r') as file:
phys_regressors = pd.read_csv(phys_regressors, delim_whitespace=True, header=None)
return phys_regressors
physio_correction = Node(Function(input_names=["script_mat", "subject", "session", "task", "table", "derivatives_dir"],
output_names=["phys_regressors"],
function= physio_toolbox),
name='physio_toolbox')
physio_correction.inputs.script_mat = script_mat_file
physio_correction.inputs.table = table
physio_correction.inputs.derivatives_dir = derivatives_dir
physio_correction.inputs.subject = "ZI004"
physio_correction.inputs.session = "03"
physio_correction.inputs.task = "faces"
denoising_node = Node(Function(input_names = ["func_path", "mask_path", "phys_regressors", "bids_dir"],
output_names = ["denoise_path"],
function = denoise_img),
name = "denoising")
denoising_node.inputs.func_path = func_path
denoising_node.inputs.mask_path = mask_path
denoising_node.inputs.bids_dir = bids_dir
# Get Subject Info - get subject specific condition information
getsubjectinfo = Node(Function(input_names=['subject', 'session', "task", "rawdata_dir"],
output_names=['subject_info'],
function=subjectinfo),
name='getsubjectinfo')
getsubjectinfo.inputs.subject = "ZI004"
getsubjectinfo.inputs.session = "03"
getsubjectinfo.inputs.task = "faces"
getsubjectinfo.inputs.rawdata_dir = rawdata_dir
from nipype.algorithms.misc import Gunzip
gunzip = Node(Gunzip(), name='gunzip')
gunzip.inputs.in_file = "/data/Stepan_Tikhomirov/EPIsoDE/derivatives/fmriprep-preprocess/fmriprep/sub-ZI004/ses-03/func/sub-ZI004_ses-03_task-faces_run-1_space-MNI152NLin6Asym_res-2_desc-preproc_bold.nii.gz"
from nipype.interfaces.spm import Smooth
smooth = Node(Smooth(), name = "smooth")
smooth.inputs.fwhm = [4, 4, 4]
from nipype.algorithms.modelgen import SpecifySPMModel, SpecifyModel
from nipype.interfaces.spm import Level1Design, EstimateModel, EstimateContrast
modelspec = Node(SpecifySPMModel(concatenate_runs=False,
input_units='secs',
output_units='secs',
time_repetition= 2,
high_pass_filter_cutoff=128),
name="modelspec")
level1design = Node(Level1Design(bases={'hrf': {'derivs': [1, 0]}},
timing_units='secs',
interscan_interval= 2,
model_serial_correlations='FAST'),
name="level1design")
# EstimateModel - estimate the parameters of the model
level1estimate = Node(EstimateModel(estimation_method={'Classical': 1}),
name="level1estimate")
# EstimateContrast - estimates contrasts
level1conest = Node(EstimateContrast(contrasts= contrast), name="level1conest")
datasink = Node(DataSink(base_directory=derivatives_dir,
container= "ZI004"),
name="datasink")
workflow1 = Workflow(name = "workflow1")
workflow1.connect(physio_correction, "phys_regressors", denoising_node, "phys_regressors")
workflow1.connect(getsubjectinfo, 'subject_info', modelspec, "subject_info")
workflow1.connect(denoising_node, "denoise_path", modelspec, "functional_runs")
workflow1.connect(modelspec, "session_info", level1design, "session_info")
workflow1.connect(level1design, "spm_mat_file", level1estimate, "spm_mat_file")
workflow1.connect([(level1estimate, level1conest, [('spm_mat_file',
'spm_mat_file'),
('beta_images',
'beta_images'),
('residual_image',
'residual_image')])])
workflow1.connect([(level1conest, datasink, [('spm_mat_file', '1stLevel.spm_mat'),
('spmT_images', '1stLevel.spmT'),
('con_images', '1stLevel.con_images'),
])])
workflow1.run()
I tried to do it with and without smoothing. Also i tried to denoise my data using motion regressors calculated by fmriprep. But when model esimate node is running I get this error :
“Error using spm_est_non_sphericity Please check your data: There are no significant voxels.”
This error occurred all the time: i tried to set detrend and standard to False, changed different parameters in first level analysis, nothing changed. though the denoised timeseries looks okay.
When i set model_serial_correlations to “none” in level1design node, i got another error :
Failed ‘Model estimation’
Error using spm_spm
Please check your data: There are no inmask voxels.
I used the mask created by fmriprep. and it also looks fine.
The last thing I’d like to underline is that when i run first level analysis without denoising, or by adding regressors to subject_info, then everything works. But I would like to try subtracting regressors before GLM.
So, what can be the reason of this error?
Thank you