Summary of what happened:
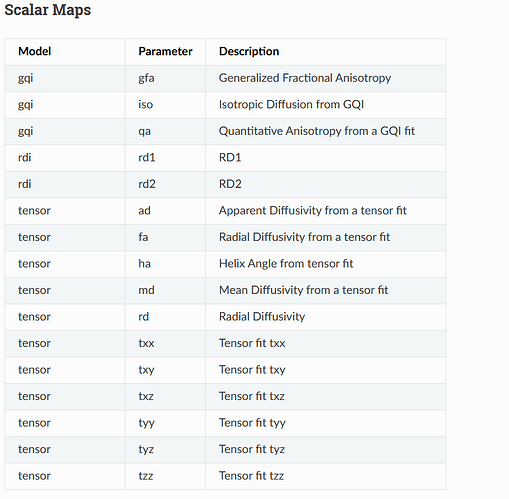
I ran qsirecon for dsi_studio_gqi on HPC for 1 subject and received no errors and received outputs. Am I missing outputs from this? I am not sure how many I should have, looking at the read the docs. Should I have all of these outputs?
My outputs in derivatives folder sub-01 dwi directory include:
(base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev$ cd qsirecon_dev_dsi_studio_gqi/
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsirecon_dev_dsi_studio_gqi$ ls
atlases dataset_description.json derivatives logs sub-01
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsirecon_dev_dsi_studio_gqi$ cd sub-01/
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsirecon_dev_dsi_studio_gqi/sub-01$ ls
dwi log
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsirecon_dev_dsi_studio_gqi/sub-01$ cd dwi/
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsirecon_dev_dsi_studio_gqi/sub-01/dwi$ ls
sub-01_run-01_space-ACPC_seg-4S256Parcels_dseg.mif.gz sub-01_run-01_space-ACPC_seg-4S256Parcels_dseg.nii.gz sub-01_run-01_space-ACPC_seg-4S256Parcels_dseg.txt
My qsiprep outputs being pulled are as follows:
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsiprep_dev_v1.0.1$ ls
dataset_description.json sub-04 sub-09 sub-13 sub-17 sub-21 sub-25 sub-31 sub-35 sub-39 sub-43 sub-47
logs sub-04.html sub-09.html sub-13.html sub-17.html sub-21.html sub-25.html sub-31.html sub-35.html sub-39.html sub-43.html sub-47.html
sub-01 sub-05 sub-10 sub-14 sub-18 sub-22 sub-26 sub-32 sub-36 sub-40 sub-44 sub-48
sub-01.html sub-05.html sub-10.html sub-14.html sub-18.html sub-22.html sub-26.html sub-32.html sub-36.html sub-40.html sub-44.html sub-48.html
sub-02 sub-06 sub-11 sub-15 sub-19 sub-23 sub-29 sub-33 sub-37 sub-41 sub-45
sub-02.html sub-06.html sub-11.html sub-15.html sub-19.html sub-23.html sub-29.html sub-33.html sub-37.html sub-41.html sub-45.html
sub-03 sub-08 sub-12 sub-16 sub-20 sub-24 sub-30 sub-34 sub-38 sub-42 sub-46
sub-03.html sub-08.html sub-12.html sub-16.html sub-20.html sub-24.html sub-30.html sub-34.html sub-38.html sub-42.html sub-46.html
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsiprep_dev_v1.0.1$ cd sub-01
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsiprep_dev_v1.0.1/sub-01$ ls
anat dwi figures log
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsiprep_dev_v1.0.1/sub-01$ cd dwi
(puma) (base) xkg@/xdisk/karpjf/xkg/derivatives/darpa_dev/qsiprep_dev_v1.0.1/sub-01/dwi$ ls
sub-01_run-01_desc-confounds_timeseries.tsv sub-01_run-01_space-ACPC_desc-preproc_dwi.bval sub-01_run-01_space-ACPC_dwiref.nii.gz
sub-01_run-01_space-ACPC_desc-brain_mask.nii.gz sub-01_run-01_space-ACPC_desc-preproc_dwi.bvec sub-01_run-01_space-ACPC_model-eddy_stat-cnr_dwimap.json
sub-01_run-01_space-ACPC_desc-image_qc.tsv sub-01_run-01_space-ACPC_desc-preproc_dwi.json sub-01_run-01_space-ACPC_model-eddy_stat-cnr_dwimap.nii.gz
sub-01_run-01_space-ACPC_desc-preproc_dwi.b sub-01_run-01_space-ACPC_desc-preproc_dwi.nii.gz
sub-01_run-01_space-ACPC_desc-preproc_dwi.b_table.txt sub-01_run-01_space-ACPC_desc-slice_qc.json
Command Used
#!/bin/bash
############################################################
############# BEGIN SLURM CONFIGURATION ######################
#SBATCH --partition=standard
#SBATCH --time=22:00:00
#SBATCH --nodes=2 --ntasks=24
#SBATCH --job-name=qsirecon
#SBATCH --account=karpjf
#SBATCH --mail-type=ALL
#SBATCH --mail-user=xkg@arizona.edu
#SBATCH --array=0
############################################################
############## BEGIN SCRIPT TO RUN #########################
# ENVIRONMENT VARIABLES
export SIF=/contrib/singularity/shared/neuroimaging
export APP=${SIF}/qsirecon_v1.1.0.sif
# Set main project directory
export MRIS=/xdisk/karpjf/xkg
# Point to the preprocessed data and freesurfer outputs
export APP_PREP_DERIV_DIR=${MRIS}/derivatives/darpa_dev/qsiprep_dev_v1.0.1
export FREESURFER_DIR=${MRIS}/derivatives/darpa_dev/fmriprep_dev_v24.1.1/sourcedata/freesurfer
export LICENSE=/groups/dkp/neuroimaging/license.txt
# Define the recon spec for this workflow
export RECON_SPEC=dsi_studio_gqi
# Define output and work directories
export APP_DERIV_DIR=${MRIS}/derivatives/darpa_dev/qsirecon_dev_dsi_studio_gqi
export WORK_DIR=${MRIS}/work/darpa_work/${RECON_SPEC}
# TEMPLATEFLOW setup
export TEMPLATEFLOW_HOME=/groups/dkp/neuroimaging/templateflow
export APPTAINTERENV_TEMPLATEFLOW_HOME=/opt/templateflow
# SUBJECT ARGUMENT
# Subject=${sub}
# Get subject ID from list using SLURM array index
Subject=$(sed -n "$((SLURM_ARRAY_TASK_ID + 1))p" /xdisk/karpjf/xkg/scripts/darpa_subjects_clean.txt)
if [ -z "${Subject}" ]; then
echo "ERROR: Subject ID could not be determined. Please check SLURM_ARRAY_TASK_ID and subject list."
exit 1
fi
if [ ! -d ${APP_DERIV_DIR} ]; then
mkdir -p ${APP_DERIV_DIR}
fi
# Create necessary directories if not present
if [ ! -d ${APP_DERIV_DIR} ]; then
mkdir -p ${APP_DERIV_DIR}
fi
if [ ! -d ${WORK_DIR} ]; then
mkdir -p ${WORK_DIR}
fi
# Run qsirecon with apptainer
apptainer run \
--containall --writable-tmpfs \
--bind ${TEMPLATEFLOW_HOME}:${APPTAINTERENV_TEMPLATEFLOW_HOME} \
--bind ${MRIS} \
--bind "${LICENSE}:/license.txt" \
${APP} \
${APP_PREP_DERIV_DIR} \
${APP_DERIV_DIR} \
participant \
--participant-label ${Subject} \
--stop-on-first-crash \
--recon-spec ${RECON_SPEC} \
--output-resolution 1.0 \
--input-type qsiprep \
--fs-license-file /license.txt \
--fs-subjects-dir "${FREESURFER_DIR}" \
-w "${WORK_DIR}" \
--nprocs 24 \
--omp-nthreads 8 \
--atlases 4S256Parcels \
-v -v
# Echo the command for logging
echo ""
echo "this is the command that was run:"
echo "apptainer run --containall --writable-tmpfs --bind ${TEMPLATEFLOW_HOME}:${APPTAINTERENV_TEMPLATEFLOW_HOME} --bind ${MRIS} --bind \"${LICENSE}:/license.txt\" ${APP} ${APP_PREP_DERIV_DIR} ${APP_DERIV_DIR} participant --participant-label ${Subject} --stop-on-first-crash --recon-spec ${RECON_SPEC} --output-resolution 1.0 --input-type qsiprep --fs-license-file /license.txt --fs-subjects-dir \"${FREESURFER_DIR}\" -w \"${WORK_DIR}\" --nprocs 24 --omp-nthreads 8 --atlases 4S256Parcels -v -v"
# Rename SLURM output
mv slurm-${SLURM_JOB_ID}_${SLURM_ARRAY_TASK_ID}.out ${MRIS}/slurm.out/slurm_qsirecon-${SLURM_JOB_ID}_${SLURM_ARRAY_TASK_ID}_${RECON_SPEC}.txt
Version:
1.1.0
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer on HPC
Data formatted according to:
BIDS