Summary of what happened:
Hello Neurostars, I am having trouble with SDC in fmriprep using field maps, please help!
I am using fmriprep to preprocess resting state and task based fmri scans, with a fieldmap, using the CMRR mb sequence. the images are getting considerably more distorted following SDC in fmriprep. I believe I am having a similar issue to this thread, but the posters did not seem to reach a conclusion.
I am using fmriprep version 23.1.04 on a high performance computing cluster, using singularity (fmriprep_23.1.04-001.sif) and slurm (the batch submission system).
I converted the raw dicoms to nifti and put them into bids format with heudiconv. I then checked the bids compliance with the bids validator, and only received minor warnings (readme file was small).
I tried using the flag --use-syn-sdc, but got the same result.
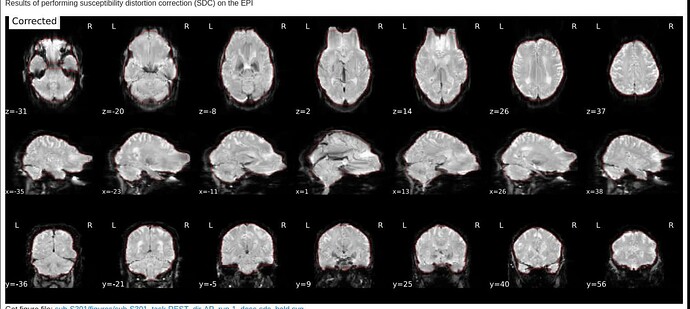
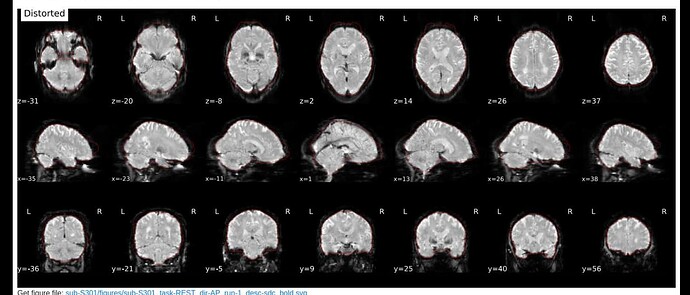
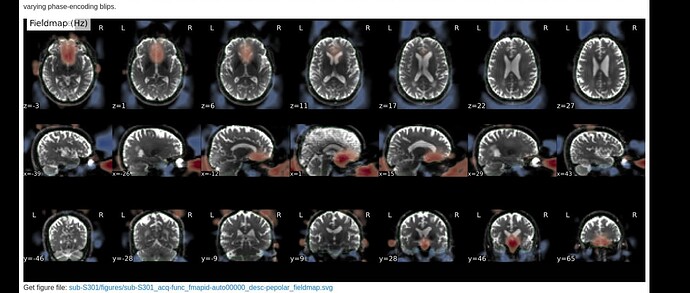
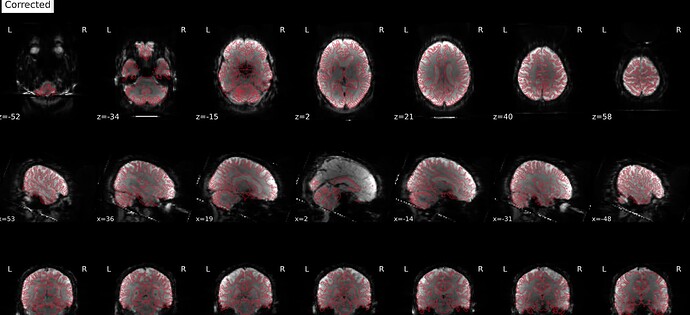
Here are images from the fmriprep html output.
Corrected:
Distorted:
and the fieldmaps:
Command used (and if a helper script was used, a link to the helper script or the command generated):
This is the code I used to run the fmriprep.sif image:
singularity run -B ${SINGULARITYENV_TEMPLATEFLOW_HOME:-$SCRATCH/software/templateflow}:/templateflow --cleanenv --home $SCRATCH -B $SCRATCH:/work $SCRATCH/software/fmriprep_23.1.4.sif ${bidsdir} ../fmriprep participant --participant-label ${subject} --fs-license-file $FS_LICENSE --me-output-echos --me-t2s-fit-method curvefit --nthreads 16 --omp-nthreads 16
Version:
The software I am using (all as singularity images)
fmriprep version 23.1.04
heudiconv version 1.0.1
bidsva.idator version 1.14.0
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity on an HPC that uses Debian as the OS and Slurm for batch submission
Data formatted according to a validatable standard? Please provide the output of the validator:
Here are my bids validator warnings (most seem benign to me):
bids-validator@1.14.0
(node:122560) Warning: Closing directory handle on garbage collection
(Use `node --trace-warnings ...` to show where the warning was created)
e[33m1: [WARN] Not all subjects contain the same files. Each subject should contain the same number of files with the same naming unless some files are known to be missing. (code: 38 - INCONSISTENT_SUBJECTS)e[39m
e[33m2: [WARN] Tabular file contains custom columns not described in a data dictionary (code: 82 - CUSTOM_COLUMN_WITHOUT_DESCRIPTION)e[39m
./sub-S301/func/sub-S301_task-REST_dir-AP_run-1_events.tsv
e[33m3: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL)e[39m
./README
Screenshots / relevant information:
Any inclination as to what might be happening? Any and all help is very much appreciated. I hope I have been adequately detailed in this post about my issue. However, I am happy to provide any further information as needed.
Thanks,
Jack