Hi,
I am using fMRIPrep (20.2.7) in a Singularity (2.6.1) container on our cluster. I get an error when trying to do this in the brain_extraction_wf - it reports a missing results file. The logs are here:
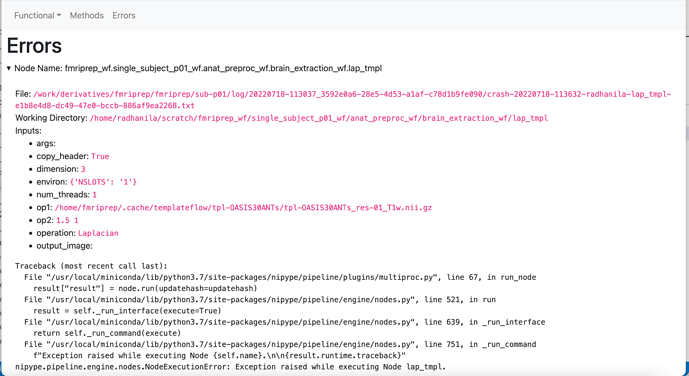
Node: fmriprep_wf.single_subject_p01_wf.anat_preproc_wf.brain_extraction_wf.lap_tmpl
Working directory: /home/radhanila/scratch/fmriprep_wf/single_subject_p01_wf/anat_preproc_wf/brain_extraction_wf/lap_tmpl
Node inputs:
args = <undefined>
copy_header = True
dimension = 3
environ = {'NSLOTS': '1'}
num_threads = 1
op1 = /home/fmriprep/.cache/templateflow/tpl-OASIS30ANTs/tpl-OASIS30ANTs_res-01_T1w.nii.gz
op2 = 1.5 1
operation = Laplacian
output_image = <undefined>
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py", line 521, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py", line 639, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py", line 751, in _run_command
f"Exception raised while executing Node {self.name}.\n\n{result.runtime.traceback}"
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node lap_tmpl.
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/interfaces/base/core.py", line 454, in aggregate_outputs
setattr(outputs, key, val)
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/usr/local/miniconda/lib/python3.7/site-packages/traits/trait_handlers.py", line 172, in error
value )
traits.trait_errors.TraitError: The 'output_image' trait of an ImageMathOuputSpec instance must be a pathlike object or string representing an existing file, but a value of '/home/radhanila/scratch/fmriprep_wf/single_subject_p01_wf/anat_preproc_wf/brain_extraction_wf/lap_tmpl/tpl-OASIS30ANTs_res-01_T1w_maths.nii.gz' <class 'str'> was specified.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/interfaces/base/core.py", line 399, in run
runtime = self._post_run_hook(runtime)
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/interfaces/mixins/fixheader.py", line 127, in _post_run_hook
outputs = self.aggregate_outputs(runtime=runtime).get_traitsfree()
File "/usr/local/miniconda/lib/python3.7/site-packages/nipype/interfaces/base/core.py", line 461, in aggregate_outputs
raise FileNotFoundError(msg)
FileNotFoundError: No such file or directory '/home/radhanila/scratch/fmriprep_wf/single_subject_p01_wf/anat_preproc_wf/brain_extraction_wf/lap_tmpl/tpl-OASIS30ANTs_res-01_T1w_maths.nii.gz' for output 'output_image' of a ImageMath interface
As suggested in this thread, I checked if the necessary template files are accessible in the container using singularity shell and they are. So, it looks like it’s the output from the workflow that is missing. I am not sure how to proceed. I’d appreciate any help.
Thanks,
Radha