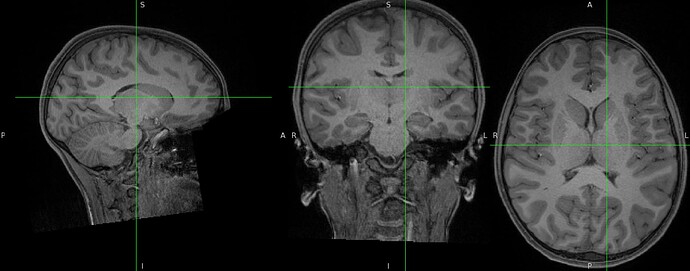
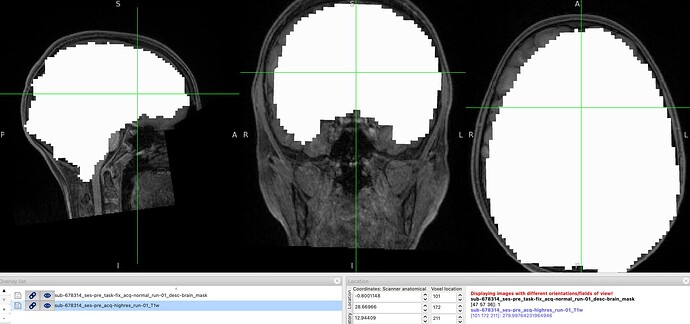
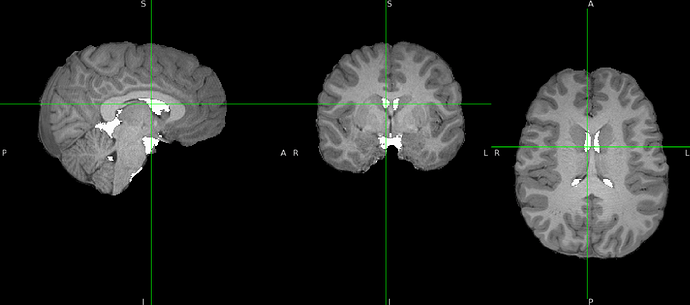
Hello everyone,
I’m using C-PAC (v1.8.7.dev1) for pre-processing rsfMRI data. I’m running the pipeline on several subjects, and I found the following error when preprocessing only one of them:
250304-16:23:57,555 nipype.workflow INFO:
[Node] Executing "build_nuisance_regressors" <CPAC.utils.interfaces.function.function.Function>
/code/CPAC/nuisance/nuisance.py:287: UserWarning: loadtxt: input contained no data: "/gpfs/scratch/martil42/ost_fix/working/pipeline_cpac-ost-fix/cpac_678314_pre/nuisance_regressors_default_163/_scan_fix_acq-normal_run-01/CerebrospinalFluid_mean/sub-678314_ses-pre_task-fix_acq-normal_run-01_bold_resample_calc_tshift_volreg_calc_maths_flirt_roistat.1D"
regressors = np.loadtxt(regressor_file)
250304-16:23:57,847 nipype.workflow INFO:
[Node] Finished "build_nuisance_regressors", elapsed time 0.051391s.
250304-16:23:57,848 nipype.workflow WARNING:
Storing result file without outputs
250304-16:23:57,849 nipype.workflow WARNING:
[Node] Error on "cpac_678314_pre.nuisance_regressors_default_163.build_nuisance_regressors" (/gpfs/scratch/martil42/ost_fix/working/pipeline_cpac-ost-fix/cpac_678314_pre/nuisance_regressors_default_163/_scan_fix_acq-normal_run-01/build_nuisance_regressors)
250304-16:23:57,851 nipype.workflow ERROR:
Node build_nuisance_regressors.a0 failed to run on host cn-0044.
250304-16:23:57,852 nipype.workflow ERROR:
Saving crash info to /gpfs/scratch/martil42/ost_fix/log/crash-20250304-162357-martil42-build_nuisance_regressors.a0-c8ac2890-1802-4fae-a5d6-7c2ae06b4e9a.txt
Traceback (most recent call last):
File "/usr/share/fsl/6.0/lib/python3.10/site-packages/nipype/pipeline/plugins/linear.py", line 47, in run
node.run(updatehash=updatehash)
File "/code/CPAC/pipeline/nipype_pipeline_engine/engine.py", line 443, in run
return super().run(updatehash)
File "/usr/share/fsl/6.0/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/usr/share/fsl/6.0/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/usr/share/fsl/6.0/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node build_nuisance_regressors.
I’m not sure where the issue might be, since the preprocessing of the same subject (but a different session) and of the remaining subjects has run smoothly, and only this one is giving errors.
Thanks in advance!!
~L