Summary of what happened:
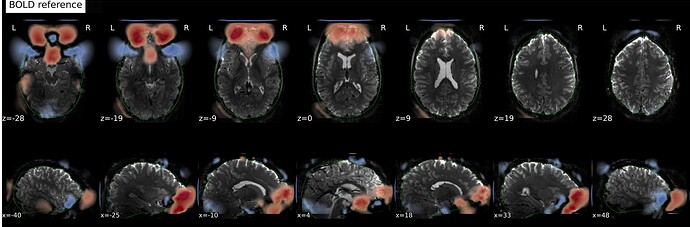
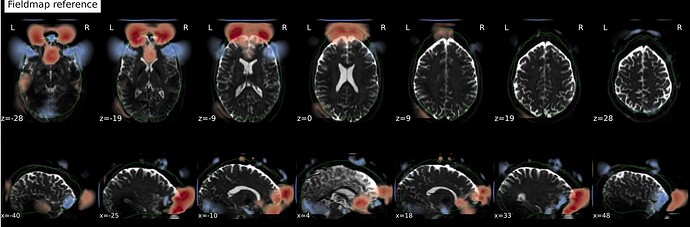
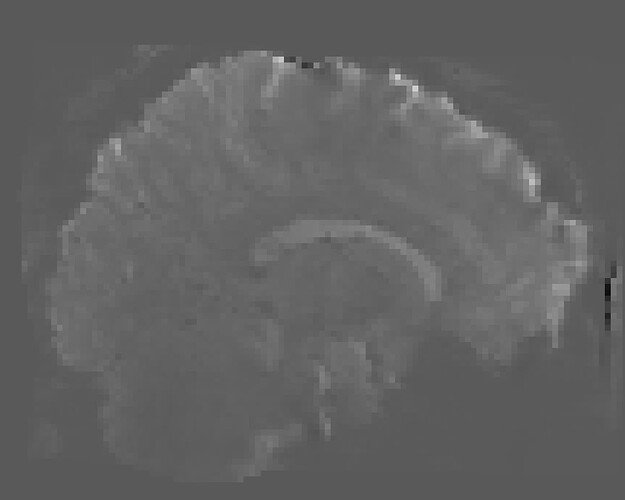
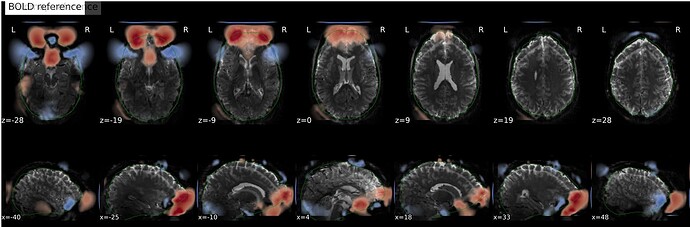
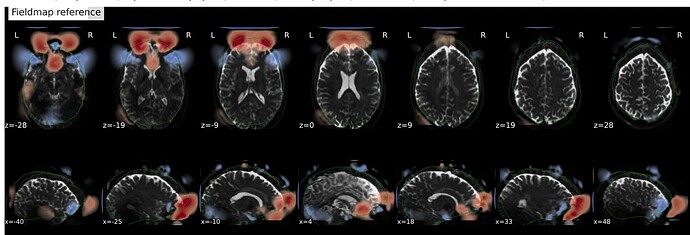
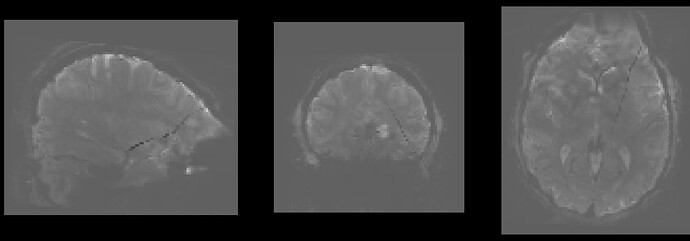
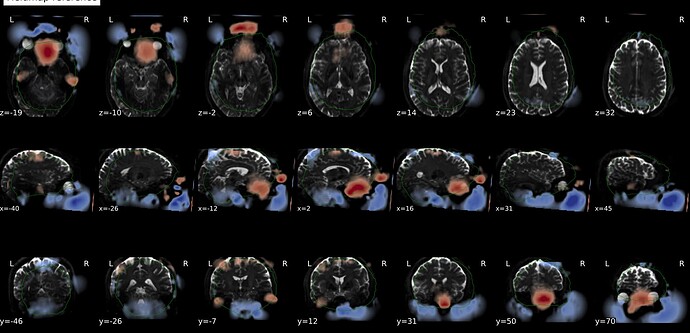
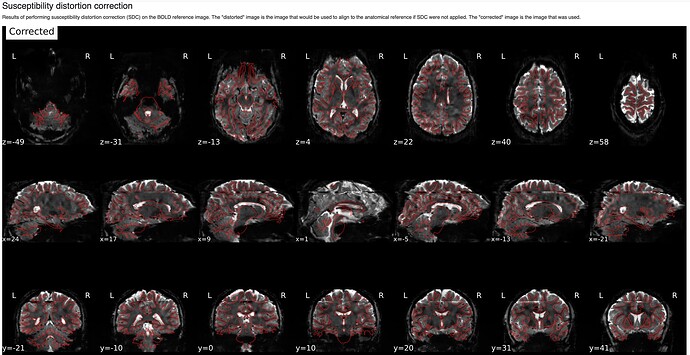
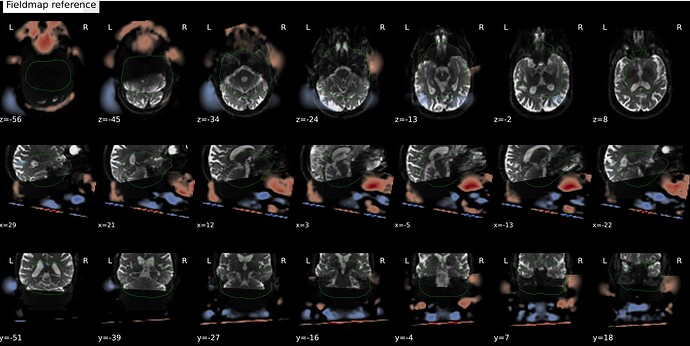
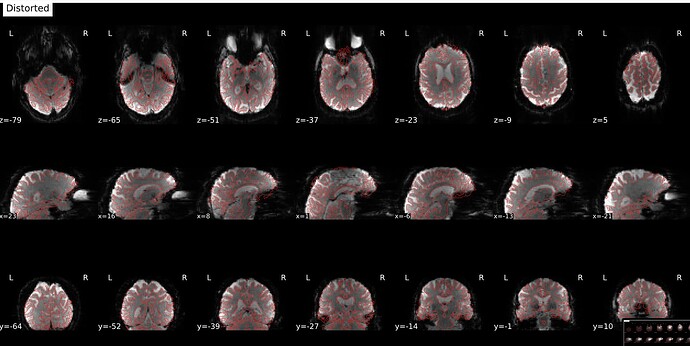
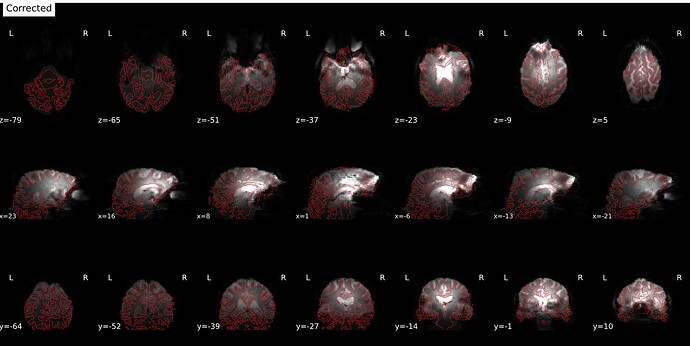
In fMRIprep, for two subjects out of a dataset of about 60 subjects, fMRIprep fails towards (I think) the end of the processing script. The output looks mostly reasonable despite the error message, except for two things that were true for both subjects: 1, There are subtle artifacts in the final data (in one case it’s just outside the brain mask, but in the other it is within the brain). 2, There seems to be a lack of alignment between the BOLD reference and the field map reference on the “Alignment between the anatomical reference of the fieldmap and the target EPI” stage of the output HTML. I’d like to figure out if there is a way to fix this issue, and if these data may be usable as-is. Note that we’re using a blip-up/blip-down PEPOLAR field map for these data.
Command used (and if a helper script was used, a link to the helper script or the command generated):
srun singularity run \
--cleanenv \
-B /project/decety \
-B /scratch/midway3/jchen28 \
/project/decety/my_images/fmriprep-24.0.0.simg \
--participant_label=5004053 \
--fs-license-file="/project/decety/freesurfer_license.txt" \
-v -v \
--nthreads 8 \
--output-spaces MNI152NLin6Asym:res-2 \
--bold2anat-dof 6 \
--fd-spike-threshold 0.9 \
--dvars-spike-threshold 3 \
--skull-strip-template OASIS30ANTs \
-w /scratch/midway3/jchen28 \
/project/decety/VAD_BIDS \
/project/decety/VAD_BIDS/derivatives/fmriprep_v24 \
participant
Version:
24.0.0
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
1: [WARN] The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. (code: 101 - README_FILE_MISSING)
Please visit https://neurostars.org/search?q=README_FILE_MISSING for existing conversations about this issue.
2: [WARN] The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered because there are no authors, which will make DOI registration from dataset metadata impossible. (code: 113 - NO_AUTHORS)
Please visit https://neurostars.org/search?q=NO_AUTHORS for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
857 Files, 318.49GB MRI
62 - Subjects
1 - Session
If you have any questions, please post on https://neurostars.org/tags/bids.
Relevant log outputs (up to 20 lines):
240801-13:36:05,95 nipype.workflow ERROR:
could not run node: fmriprep_24_0_wf.sub_5004053_wf.fmap_preproc_wf.fmap_reports_wf_auto_00000.fmap_rpt
240801-13:36:05,99 nipype.workflow INFO:
crashfile: /project/decety/VAD_BIDS/derivatives/fmriprep_v24/sub-5004053/log/20240801-115027_aa89766f-6adf-42d7-b550-7a4c4538308a/crash-20240801-115416-jchen28-fmap_rpt-5fa684b5-9739-48da-881d-bd8c613b4bbc.txt
240801-13:36:05,99 nipype.workflow INFO:
***********************************
240801-13:36:05,155 nipype.workflow CRITICAL:
fMRIPrep failed: Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node fmap_rpt.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._post_run_hook(runtime)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/mixins/reporting.py", line 50, in _post_run_hook
self._generate_report()
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/sdcflows/interfaces/reportlets.py", line 126, in _generate_report
plot_registration(
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/sdcflows/viz/utils.py", line 87, in plot_registration
svg = extract_svg(display, compress=compress)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/niworkflows/viz/utils.py", line 140, in extract_svg
image_svg = svg2str(display_object, dpi)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/niworkflows/viz/utils.py", line 132, in svg2str
display_object.frame_axes.figure.savefig(
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/figure.py", line 3390, in savefig
self.canvas.print_figure(fname, **kwargs)
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/backend_bases.py", line 2193, in print_figure
result = print_method(
^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/backend_bases.py", line 2043, in <lambda>
print_method = functools.wraps(meth)(lambda *args, **kwargs: meth(
^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/backends/backend_svg.py", line 1339, in print_svg
self.figure.draw(renderer)
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/artist.py", line 95, in draw_wrapper
result = draw(artist, renderer, *args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/artist.py", line 72, in draw_wrapper
return draw(artist, renderer)
^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/figure.py", line 3143, in draw
artists = self._get_draw_artists(renderer)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/matplotlib/figure.py", line 231, in _get_draw_artists
ax.apply_aspect(locator(ax, renderer) if locator else None)
^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nilearn/plotting/displays/_slicers.py", line 1579, in _locator
[[left_dict[axes], y0], [left_dict[axes] + width_dict[axes], y1]]
~~~~~~~~~^^^^^^
KeyError: <Axes: >
Screenshots / relevant information:
First affected subject:
fMRIprep output:
Preprocessed BOLD image (see black artifact at the front of the brain):
Second affected subject:
fMRIprep output
Preprocessed BOLD image (see dark line cutting through brain):