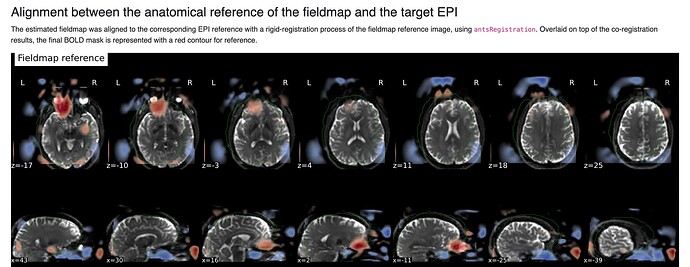
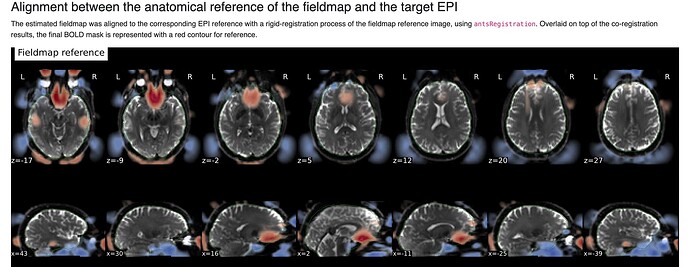
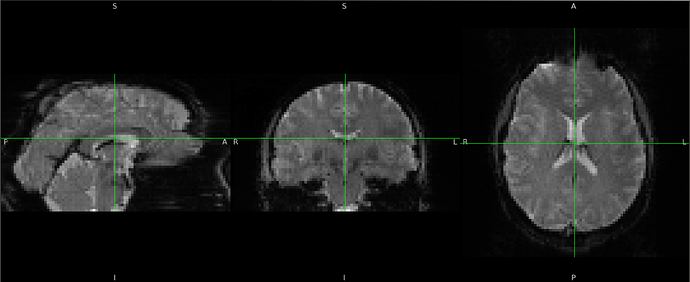
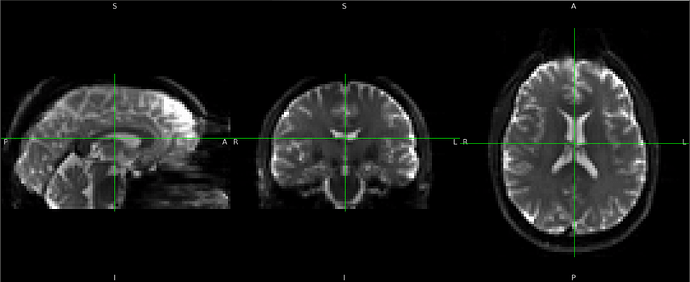
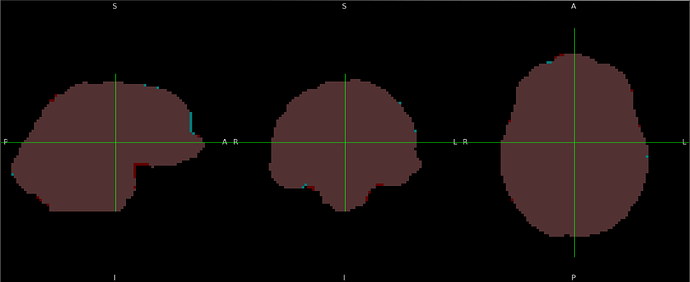
Summary of what happened:
I’m testing the newest version of fMRIPrep on a small dataset, 1 participant scanned on 9 scanners. For one of the sessions, the EPI ↔ functional registration is quite bad.
In that run, an anatomical mask was provided through the --derivatives argument (it’s a mask calculated by synthstrip and is very good). However, when I don’t provide the mask, the registration looks fine.
(The exclusion of cerebellum is real and not an artifact of figure cropping)
Note that the registration ends up bad when using or not using synthstrip’s --no-csf flag.
Are there gotcha’s when providing pre-specified masks? For example, when providing anatomical masks, do I also need to provide masks for the functionals (and/or fmaps)? Or, do I need to ensure that the mask covers or doesn’t cover certain structures (brainstem, cerebellum, etc)?
(FWIW, if this seems like a possible bug, I’d be happy/motivated to dig around)
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker run \
--rm -it \
-v $PWD:$PWD \
nipreps/fmriprep:24.0.1 \
--notrack \
--fs-license-file $PWD/license \
--work-dir $PWD/work2 \
--task-id rest \
--derivatives synthstrip=$PWD/sourcedata/synthstrip \
--dummy-scans 15 \
$PWD/rawdata \
$PWD/derivatives/fmriprep-synthstrip \
participant
vs
docker run \
--rm -it \
-v $PWD:$PWD \
nipreps/fmriprep:24.0.1 \
--notrack \
--fs-license-file $PWD/license \
--dummy-scans 15 \
--work-dir $PWD/work \
--task-id rest \
$PWD/rawdata \
$PWD/derivatives \
participant
Version:
24.0.1
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.14.6
(node:9) Warning: Closing directory handle on garbage collection
(Use `node --trace-warnings ...` to show where the warning was created)
1: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL)
./README
Please visit https://neurostars.org/search?q=README_FILE_SMALL for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
18 Files, 771.33MB cuff MRI
1 - Subject rest
1 - Session
If you have any questions, please post on https://neurostars.org/tags/bids.
Relevant log outputs (up to 20 lines):
240905-21:54:38,271 nipype.workflow IMPORTANT:
Building fMRIPrep's workflow:
* BIDS dataset path: /Users/psadil/Desktop/NStravel2/rawdata.
* Participant list: ['travel2'].
* Run identifier: 20240905-215427_aff96021-2fa0-443f-8faf-7280522b1da7.
* Output spaces: MNI152NLin2009cAsym:res-native.
* Searching for derivatives: [PosixPath('/Users/psadil/Desktop/NStravel2/sourcedata/synthstrip')].
* Pre-run FreeSurfer's SUBJECTS_DIR: /Users/psadil/Desktop/NStravel2/derivatives/fmriprep-synthstrip/sourcedata/freesurfer.
240905-21:54:38,814 nipype.workflow INFO:
ANAT Stage 1: Adding template workflow
240905-21:54:39,171 nipype.workflow INFO:
ANAT Found brain mask
240905-21:54:39,171 nipype.workflow INFO:
ANAT Skipping skull-strip, INU-correction only
240905-21:54:39,186 nipype.workflow INFO:
ANAT Stage 3: Preparing segmentation workflow
240905-21:54:39,190 nipype.workflow INFO:
ANAT Stage 4: Preparing normalization workflow for ['MNI152NLin2009cAsym']
240905-21:54:39,199 nipype.workflow INFO:
ANAT Stage 5: Preparing surface reconstruction workflow
240905-21:54:39,215 nipype.workflow INFO:
ANAT Found brain mask - skipping Stage 6
Screenshots / relevant information:
See above