@emdupre @ChrisGorgolewski
Hi,
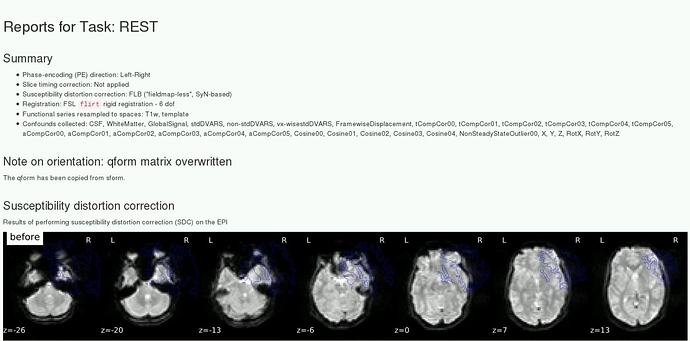
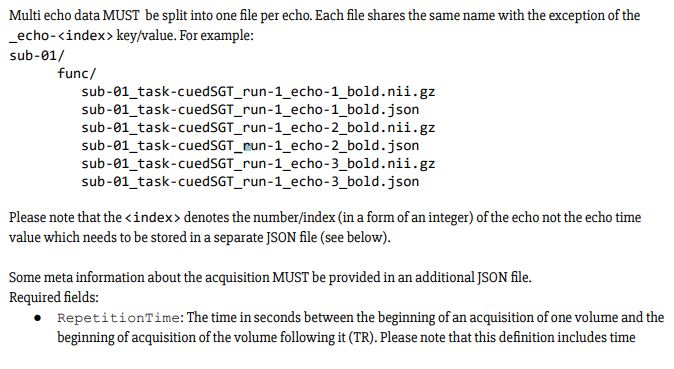
This took longer than expected. However, I have managed to run fMRIPrep v1.2.5 on ME-data, and slice time correction has been applied! Yay!
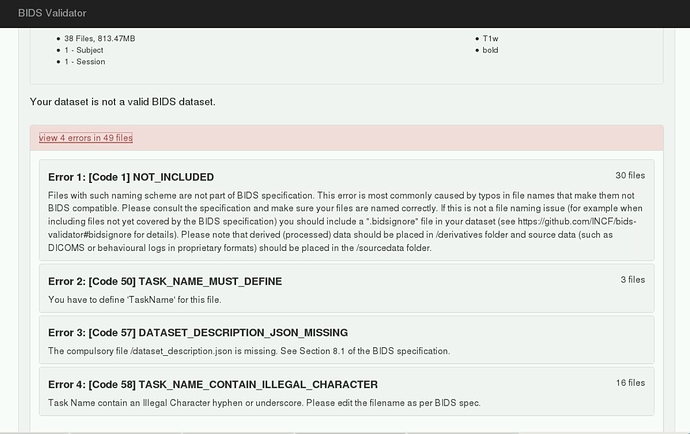
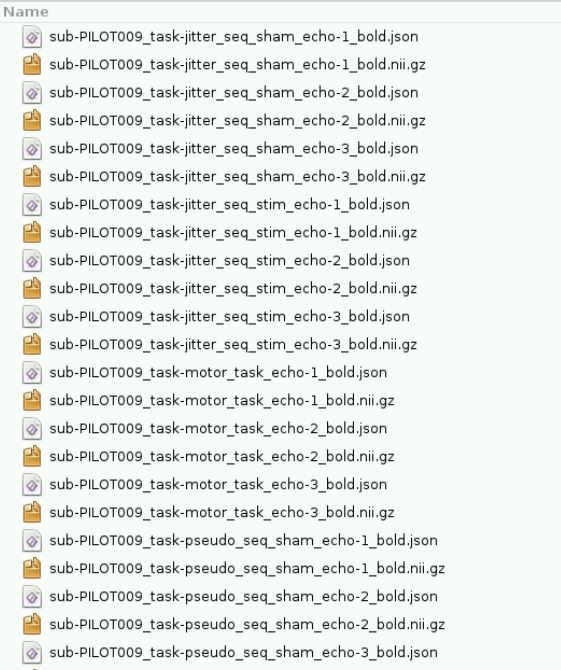
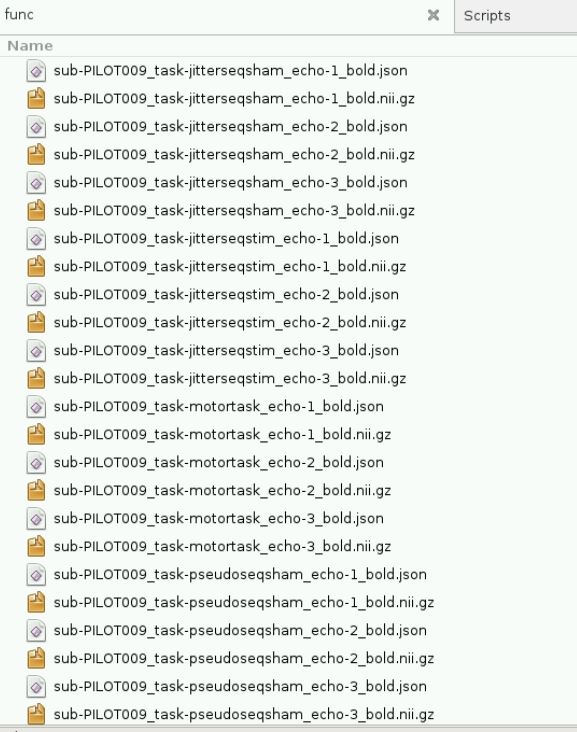
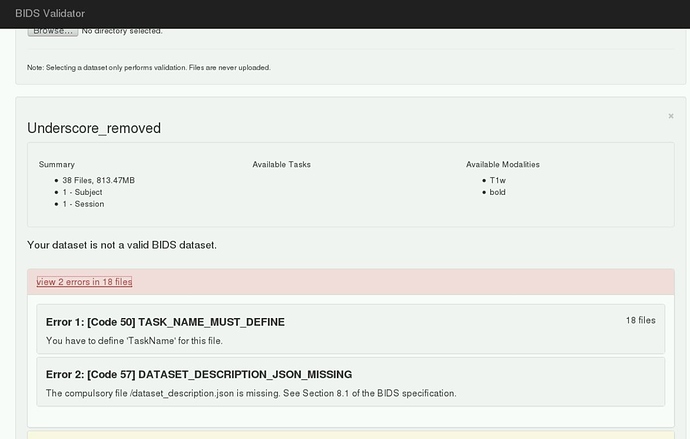
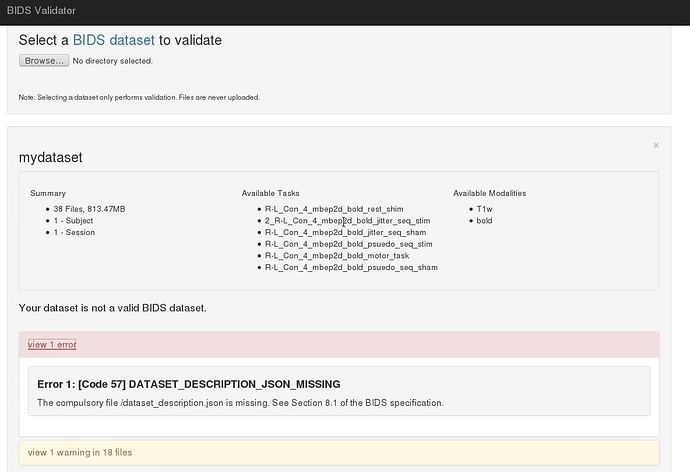
However, initially I ran into memory issues despite increasing the memory to 12 gigs. I think this was when fMRIPrep tried running the BIDS validation step, becauase only when I skipped this step was fMRIPrep able to process my data. Please see error below:
bash fMRI_prep_multi_echo_v2_TMSfMRI.sh
---------------------------
----- 1 sub-PILOT010 -----
---------------------------
Making sure the input data is BIDS compliant (warnings can be ignored in most cases).
<--- Last few GCs --->
[7852:0x367af80] 24038 ms: Mark-sweep 1392.2 (1398.5) -> 1392.2 (1397.5) MB, 5.0 / 0.0 ms (average mu = 0.985, current mu = 0.002) last resort GC in old space requested
[7852:0x367af80] 24043 ms: Mark-sweep 1392.2 (1397.5) -> 1392.2 (1397.5) MB, 4.8 / 0.0 ms (average mu = 0.974, current mu = 0.002) last resort GC in old space requested
<--- JS stacktrace --->
==== JS stack trace =========================================
0: ExitFrame [pc: 0x2beb882dc01d]
Security context: 0x11a55fc63249 <JSObject>
1: stringSlice(aka stringSlice) [0x1bc12ce1b9d1] [buffer.js:595] [bytecode=0x1bc12ce557b1 offset=91](this=0x198a1cc826f1 <undefined>,buf=0x2b61d02fe9f9 <Uint8Array map = 0x85b17250599>,encoding=0x05a2a02a0bf1 <String[4]: utf8>,start=0,end=49250463)
2: toString [0x5a2a029b749] [buffer.js:668] [bytecode=0x1bc12ce552a1 offset=145](this=0x2b61d02fe9f9 <U...
FATAL ERROR: CALL_AND_RETRY_LAST Allocation failed - JavaScript heap out of memory
1: 0x8d20d0 node::Abort() [bids-validator]
2: 0x8d211c [bids-validator]
3: 0xb02b6e v8::Utils::ReportOOMFailure(v8::internal::Isolate*, char const*, bool) [bids-validator]
4: 0xb02da4 v8::internal::V8::FatalProcessOutOfMemory(v8::internal::Isolate*, char const*, bool) [bids-validator]
5: 0xef02e2 [bids-validator]
6: 0xeffb4f v8::internal::Heap::AllocateRawWithRetryOrFail(int, v8::internal::AllocationSpace, v8::internal::AllocationAlignment) [bids-validator]
7: 0xec7e35 [bids-validator]
8: 0xecf79b v8::internal::Factory::NewRawTwoByteString(int, v8::internal::PretenureFlag) [bids-validator]
9: 0xed0072 v8::internal::Factory::NewStringFromUtf8(v8::internal::Vector<char const>, v8::internal::PretenureFlag) [bids-validator]
10: 0xb10979 v8::String::NewFromUtf8(v8::Isolate*, char const*, v8::NewStringType, int) [bids-validator]
11: 0x994878 node::StringBytes::Encode(v8::Isolate*, char const*, unsigned long, node::encoding, v8::Local<v8::Value>*) [bids-validator]
12: 0x8edf32 [bids-validator]
13: 0xb8b32f [bids-validator]
14: 0xb8be99 v8::internal::Builtin_HandleApiCall(int, v8::internal::Object**, v8::internal::Isolate*) [bids-validator]
15: 0x2beb882dc01d
Traceback (most recent call last):
File "/usr/local/miniconda/bin/fmriprep", line 11, in <module>
sys.exit(main())
File "/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/cli/run.py", line 353, in main
validate_input_dir(exec_env, opts.bids_dir, opts.participant_label)
File "/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/cli/run.py", line 534, in validate_input_dir
subprocess.check_call(['bids-validator', bids_dir, '-c', [temp.name](http://temp.name/)])
File "/usr/local/miniconda/lib/python3.6/subprocess.py", line 291, in check_call
raise CalledProcessError(retcode, cmd)
subprocess.CalledProcessError: Command '['bids-validator', '/home/ttha0011/kg98/Thapa/TMS-fMRI_Project/Pilot_testing/MEICAvfMRIPrep/fMRIPrep/rawdata', '-c', '/tmp/tmplep73xbs']' died with <Signals.SIGABRT: 6>.
Sentry is attempting to send 1 pending error messages
Waiting up to 2.0 seconds
Press Ctrl-C to quit
----- DONE ----
P.S. happy to raise this as a separate issue in a different post.
Thank you both for your help 
Cheers,
Thapa