Hello,
I have a case that has an odd T1 segmentation from the first anat step in fMRIPrep. Does anyone know what is happening? Is there a way to fix this?
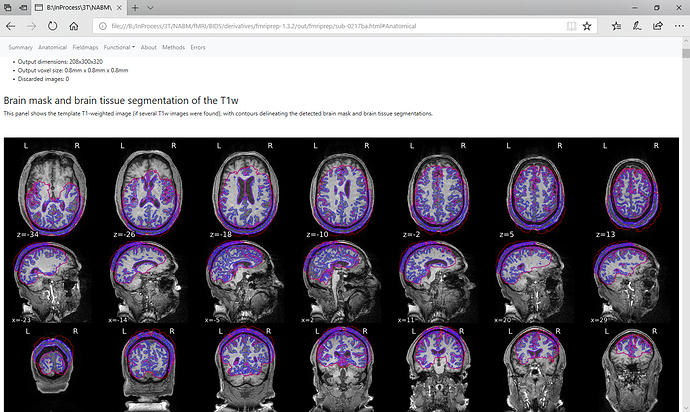
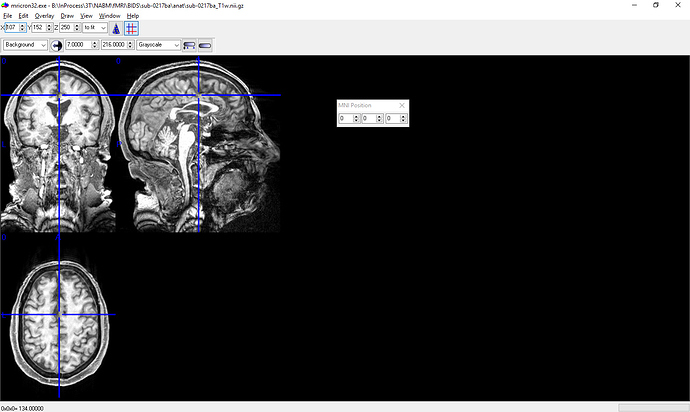
I have attached screenshots of the .nii image from MRIcroN and from fMRIPrep’s html output. The image looks fine in MRICroN.
Thanks.
The root of this problem appears to be a bad skull-stripping, perhaps due to a bad initial registration of the skull-stripping template. Could you provide your full fmriprep command?
And I wonder if you would be able to upload a defaced copy of your T1w image somewhere we could have a look at it.
Here is my command: docker run -v /opt/freesurfer/6.0.0/license.txt:/fslicense.txt:ro -v /m/InProcess/3T/NABM/fMRI/BIDS:/data:ro -v /m/InProcess/3T/NABM/fMRI/BIDS/derivatives/fmriprep-1.3.2:/work -w /work poldracklab/fmriprep:1.3.2 /data out/ participant --participant_label 0217ba --ignore slicetiming --fs-license-file /fslicense.txt
It has worked well on other participants so far. I have defaced the T1 using pydeface. Unfortunately, I am on a network that does not allow cloud/shared drives, and can’t attach to the forum. Can I email you?
Thanks.
Sure. This user @gmail will work.
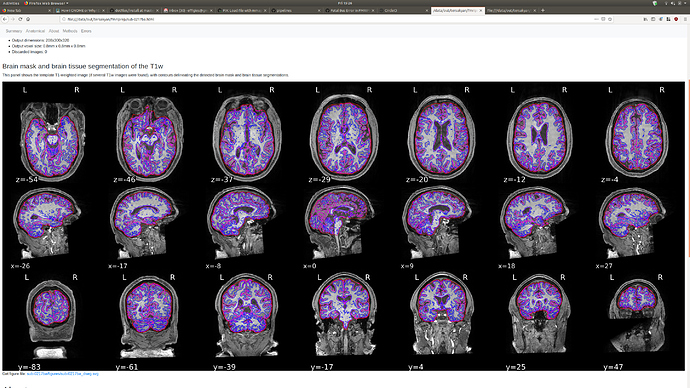
Hi @atersakyan, I actually did not have any problems running fMRIPrep on this file.
If you still have your working directory, delete the fmriprep_wf/single_subject_0217ba_wf/anat_preproc_wf/brain_extraction_wf subdirectory, and see if re-running resolves the issue. It may be that the random initialization hit an unlikely error condition.
One other difference is that I used the latest releases (1.4.1), so if you get the same result as before, you can try upgrading.
Another possibility is that defacing for some reason makes the initial registration work a bit better. You could try replacing your T1w.nii.gz files with the defaced versions.
2 Likes
Thank you, I will try those suggestions.