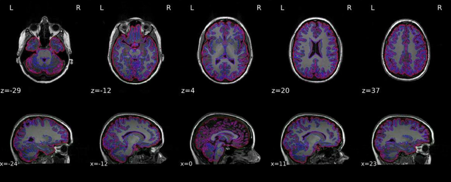
Dear all,
My output from the fmripreproc preprocessing looks very strange!
The co-registration is way off and the striatum has been moved to the frontal cortex?
Does anyone know why this might of happened? I did not use skull stripped images - would this help?
I’m attaching example Asym_desc_prerpoc_T1
Faith
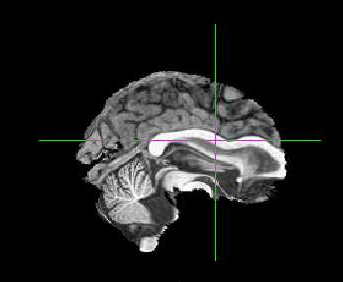
Hi @fborgan, can you post a full report? I’m afraid brain extraction is horribly failing for you.
Hi @oesteban, Yes here you go:
Summary
Subject ID: sub-021
Structural images: 1 T1-weighted
Functional series: 1
Task: rest (1 run)
Resampling targets: MNI152NLin2009cAsym, fsaverage5
FreeSurfer reconstruction: Run by fMRIPrep
Anatomical
Anatomical Conformation
Input T1w images: 1
Output orientation: RAS
Output dimensions: 196x252x197
Output voxel size: 1.2mm x 1mm x 1mm
Discarded images: 0
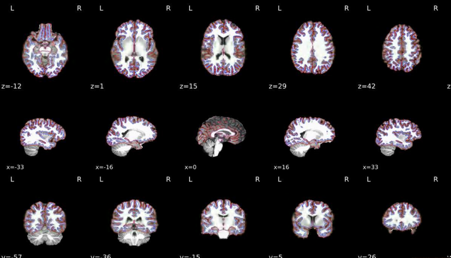
Brain mask and brain tissue segmentation of the T1w
This panel shows the template T1-weighted image (if several T1w images were found), with contours delineating the detected brain mask and brain tissue segmentations.
filename:sub-021/figures/sub-021_seg_brainmask.svg
Get figure file: sub-021/figures/sub-021_seg_brainmask.svg
T1 to MNI registration
Nonlinear mapping of the T1w image into MNI space. Hover on the panel with the mouse to transition between both spaces.
filename:sub-021/figures/sub-021_t1_2_mni.svg
Get figure file: sub-021/figures/sub-021_t1_2_mni.svg
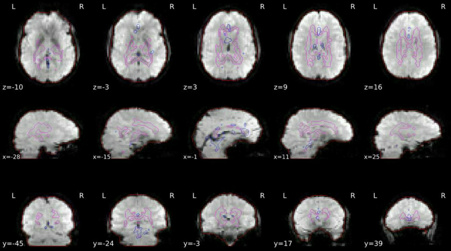
Surface reconstruction
Surfaces (white and pial) reconstructed with FreeSurfer (recon-all) overlaid on the participant’s T1w template.
filename:sub-021/figures/sub-021_reconall.svg
Get figure file: sub-021/figures/sub-021_reconall.svg
Functional
Summary
Phase-encoding (PE) direction: MISSING - Assuming Anterior-Posterior
Slice timing correction: Applied
Susceptibility distortion correction: None
Registration: FreeSurfer bbregister (boundary-based registration, BBR) - 6 dof
Functional series resampled to spaces: template, fsaverage5
Confounds collected: csf, white_matter, global_signal, std_dvars, dvars, framewise_displacement, t_comp_cor_00, t_comp_cor_01, t_comp_cor_02, t_comp_cor_03, t_comp_cor_04, t_comp_cor_05, a_comp_cor_00, a_comp_cor_01, a_comp_cor_02, a_comp_cor_03, a_comp_cor_04, a_comp_cor_05, cosine00, cosine01, cosine02, cosine03, trans_x, trans_y, trans_z, rot_x, rot_y, rot_z
Note on orientation: qform matrix overwritten
The qform has been copied from sform.
ROIs in BOLD space
Brain mask calculated on the BOLD signal (red contour), along with the masks used for a/tCompCor.
The aCompCor mask (magenta contour) is a conservative CSF and white-matter mask for extracting physiological and movement confounds.
The fCompCor mask (blue contour) contains the top 5% most variable voxels within a heavily-eroded brain-mask.
filename:sub-021/figures/sub-021_task-rest_rois.svg
Get figure file: sub-021/figures/sub-021_task-rest_rois.svg
EPI to T1 registration
bbregister was used to generate transformations from EPI-space to T1w-space
filename:sub-021/figures/sub-021_task-rest_bbregister.svg
Get figure file: sub-021/figures/sub-021_task-rest_bbregister.svg
BOLD Summary
Summary statistics are plotted, which may reveal trends or artifacts in the BOLD data. Global signals calculated within the whole-brain (GS), within the white-matter (WM) and within cerebro-spinal fluid (CSF) show the mean BOLD signal in their corresponding masks. DVARS and FD show the standardized DVARS and framewise-displacement measures for each time point.
A carpet plot shows the time series for all voxels within the brain mask. Voxels are grouped into cortical (blue), and subcortical (orange) gray matter, cerebellum (green) and white matter and CSF (red), indicated by the color map on the left-hand side.
filename:sub-021/figures/sub-021_task-rest_carpetplot.svg
Get figure file: sub-021/figures/sub-021_task-rest_carpetplot.svg
About
FMRIPrep version: 1.2.6-1
FMRIPrep command: /usr/local/miniconda/bin/fmriprep /home/k1204852/fmri_data/data/resting/my_dataset/ /home/k1204852/fmri_data/data/resting/output_folder/ participant --participant-label sub-021 --task-id rest --fs-license-file /home/k1204852/license.txt
Date preprocessed: 2019-07-08 12:18:15 +0100
Methods
We kindly ask to report results preprocessed with fMRIPrep using the following boilerplate
HTML
Markdown
LaTeX
Results included in this manuscript come from preprocessing performed using fMRIPprep 1.2.6-1 (Esteban, Markiewicz, et al. (2018); Esteban, Blair, et al. (2018); RRID:SCR_016216), which is based on Nipype 1.1.7 (Gorgolewski et al. (2011); Gorgolewski et al. (2018); RRID:SCR_002502).
Anatomical data preprocessing
The T1-weighted (T1w) image was corrected for intensity non-uniformity (INU) using N4BiasFieldCorrection (Tustison et al. 2010, ANTs 2.2.0), and used as T1w-reference throughout the workflow. The T1w-reference was then skull-stripped using antsBrainExtraction.sh (ANTs 2.2.0), using OASIS as target template. Brain surfaces were reconstructed using recon-all (FreeSurfer 6.0.1, RRID:SCR_001847, Dale, Fischl, and Sereno 1999), and the brain mask estimated previously was refined with a custom variation of the method to reconcile ANTs-derived and FreeSurfer-derived segmentations of the cortical gray-matter of Mindboggle (RRID:SCR_002438, Klein et al. 2017). Spatial normalization to the ICBM 152 Nonlinear Asymmetrical template version 2009c (Fonov et al. 2009, RRID:SCR_008796) was performed through nonlinear registration with antsRegistration (ANTs 2.2.0, RRID:SCR_004757, Avants et al. 2008), using brain-extracted versions of both T1w volume and template. Brain tissue segmentation of cerebrospinal fluid (CSF), white-matter (WM) and gray-matter (GM) was performed on the brain-extracted T1w using fast (FSL 5.0.9, RRID:SCR_002823, Zhang, Brady, and Smith 2001).
Functional data preprocessing
For each of the 1 BOLD runs found per subject (across all tasks and sessions), the following preprocessing was performed. First, a reference volume and its skull-stripped version were generated using a custom methodology of fMRIPrep. The BOLD reference was then co-registered to the T1w reference using bbregister (FreeSurfer) which implements boundary-based registration (Greve and Fischl 2009). Co-registration was configured with nine degrees of freedom to account for distortions remaining in the BOLD reference. Head-motion parameters with respect to the BOLD reference (transformation matrices, and six corresponding rotation and translation parameters) are estimated before any spatiotemporal filtering using mcflirt (FSL 5.0.9, Jenkinson et al. 2002). BOLD runs were slice-time corrected using 3dTshift from AFNI 20160207 (Cox and Hyde 1997, RRID:SCR_005927). The BOLD time-series, were resampled to surfaces on the following spaces: fsaverage5. The BOLD time-series (including slice-timing correction when applied) were resampled onto their original, native space by applying a single, composite transform to correct for head-motion and susceptibility distortions. These resampled BOLD time-series will be referred to as preprocessed BOLD in original space, or just preprocessed BOLD. The BOLD time-series were resampled to MNI152NLin2009cAsym standard space, generating a preprocessed BOLD run in MNI152NLin2009cAsym space. First, a reference volume and its skull-stripped version were generated using a custom methodology of fMRIPrep. Several confounding time-series were calculated based on the preprocessed BOLD: framewise displacement (FD), DVARS and three region-wise global signals. FD and DVARS are calculated for each functional run, both using their implementations in Nipype (following the definitions by Power et al. 2014). The three global signals are extracted within the CSF, the WM, and the whole-brain masks. Additionally, a set of physiological regressors were extracted to allow for component-based noise correction (CompCor, Behzadi et al. 2007). Principal components are estimated after high-pass filtering the preprocessed BOLD time-series (using a discrete cosine filter with 128s cut-off) for the two CompCor variants: temporal (tCompCor) and anatomical (aCompCor). Six tCompCor components are then calculated from the top 5% variable voxels within a mask covering the subcortical regions. This subcortical mask is obtained by heavily eroding the brain mask, which ensures it does not include cortical GM regions. For aCompCor, six components are calculated within the intersection of the aforementioned mask and the union of CSF and WM masks calculated in T1w space, after their projection to the native space of each functional run (using the inverse BOLD-to-T1w transformation). The head-motion estimates calculated in the correction step were also placed within the corresponding confounds file. All resamplings can be performed with a single interpolation step by composing all the pertinent transformations (i.e. head-motion transform matrices, susceptibility distortion correction when available, and co-registrations to anatomical and template spaces). Gridded (volumetric) resamplings were performed using antsApplyTransforms (ANTs), configured with Lanczos interpolation to minimize the smoothing effects of other kernels (Lanczos 1964). Non-gridded (surface) resamplings were performed using mri_vol2surf (FreeSurfer).Hi @oesteban,
Is that the report that you were referring to?
Any suggestions would be greatly appreciated!
Thanks,
Faith
Hi @fborgan,
Can you zip together the sub-021.html file and the corresponding figures under sub-021/figures/*? You will find these files under /home/k1204852/fmri_data/data/resting/output_folder/fmriprep.
Thanks very much
Hi @oesteban
Yes ofcourse. Unfortunately, I cannot upload these formats in Neurostars.
You can view these files from my googledrive account: https://drive.google.com/drive/folders/1W3U5c9sa68tRZNFa2JizUTdpabU_sBIm?usp=sharing
Thanks in advance for your help!
Faith
Hi @fborgan, this looks very much like fMRIPrep bad anatomical/T1 segmentation. Could you have a look into that issue?
Hi @oesteban,
Great, thanks.
It looks like the main advice from the thread was to delete everything and re-run the analysis. Is there anything else that you would recommend doing in addition to this?
Thanks,
Faith
I believe removing the work directory should do
Did wiping the working directory work out?
Hi @oesteban,
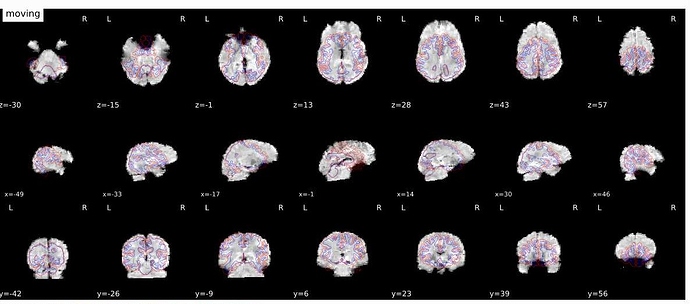
I re-ran everything but the output still looks quite odd - please see attached.
Do you think it would help to do the brain extraction first?
Thanks,
Faith
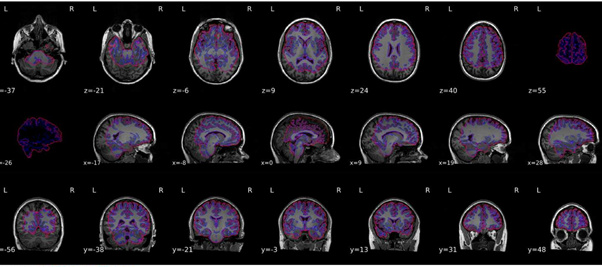
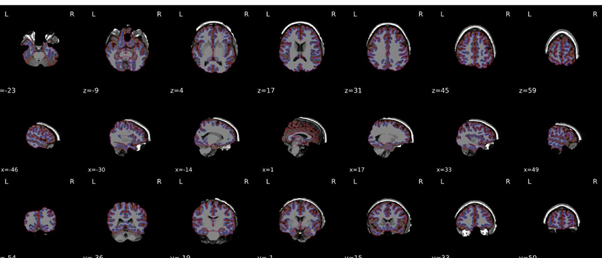
I think you are still reusing the work directory. Could you double check? Any possibility you could get us those data for us to replicate?
Hi @oesteban,
I deleted everything from the work directory and made a new output folder (output2).
Yes ofcourse - you can download the structural and functional files for 1 subject (shown above) from here: https://drive.google.com/open?id=1vuPcADqugafYKhW4DYdONEweR3fh1fLK
Thanks,
Faith
Couldn’t replicate, my reports look good (https://drive.google.com/file/d/1Sj5R6C1l8v2mZJACwJzCFdURtqzVQQnF/view?usp=sharing).
Are you trying to reuse a past FreeSurfer run? Otherwise, you’ll need to wipe your work directory (please note, I’m talking about the work directory and not about the outputs directory).
I’ve tried running it again and the output looks better.