Summary of what happened:
I have run fmriprep using two versions: 20.2.2 (with custom code to use align_epi_anat.py from afni to align bold to T1w) and 25.1.3. In the new version, I notice the bold anterior ventricles do not overlay completely onto the preprocessed T1w image. In the old version, the ventricle alignment looks perfect. I see this misalignment often in our nonmultiecho fmri data. Does anyone else notice a misalignment usually in the front ventricles in new fmriprep versions?
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv \
-B /scratch/g/jbinder/jheffernan/experiments/tdcs/bids:/data \
-B /scratch/g/jbinder/jheffernan/experiments/tdcs/derivatives/fmriprep-v25.1.3/fmriprep:/out \
-B /scratch/g/jbinder/jheffernan/experiments/tdcs/derivatives/fmriprep-v25.1.3/tmp/sub-MCW052:/work \
-B /hpc/apps/freesurfer/6.0.0/license.txt:/opt/freesurfer/license.txt \
/scratch/g/jbinder/jheffernan/experiments/tdcs/singularity_images/fmriprep-v25.1.3.sif \
/data /out participant \
--participant_label MCW052 \
--nprocs 16 \
--omp-nthreads 4 \
--output-spaces MNI152NLin2009cAsym:res-2 T1w \
--no-submm-recon \
--fs-license-file /opt/freesurfer/license.txt \
--work-dir /work \
--bold2anat-dof 6 \
--fs-no-reconall \
--ignore slicetiming
Version:
fMRIPrep 25.1.3 and fMRIPrep 20.2.2
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
Our dataset is in bids format and fMRIPrep successfully completes.
Screenshots / relevant information:
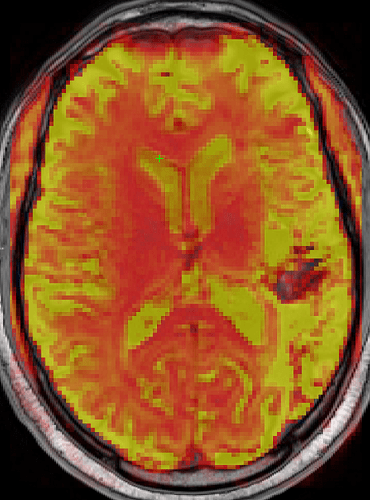
I’m not sure how bet to present the data, but here is the fMRIPrep 20.2.2 bold at 0.45 opacity in T1w space overlaid onto the preprocessed T1w image:
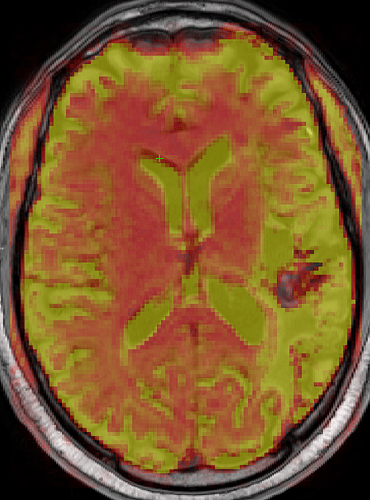
Here is the fMRIPrep 25.1.3 bold at 0.45 opacity in T1w space overlaid onto the same preprocessed T1w image:
The green crosshair is in the same location in both images. The same T1w image is used as the background in both. Notice in the second image the anterior ventricles do not complete overlap the T1w image ventricles where the crosshair is located.
Is the alignment in the second image common when preprocessing bold? I feel like there should be maximal alignment between the T1w and bold ventricles especially if we are creating a CSF regressor.