Summary of what happened:
I have been trying to get fMRIprep running on the HPC cluster I’m working with for almost two weeks now. This bug in particular I am having a hard time debugging since there isn’t really much information to go off of. If there is any way that I can provide more useful information please let me know because that would probably be helpful for myself too.
Command used (and if a helper script was used, a link to the helper script or the command generated):
module load apptainer
project=~/projects/<lab group>/<username>/bird_data_analysis
sub_num="05"
# Extract zipped BOLD data to temp directory
cp ${project}/data/raw_data/sub_${sub_num}.tar.gz $SLURM_TMPDIR/
# Places fmri_processing directory in SLURM_TMPDIR
tar -xzf $SLURM_TMPDIR/sub_${sub_num}.tar.gz -C $SLURM_TMPDIR/
# Create directories for fMRIprep to access at runtime
mkdir $SLURM_TMPDIR/work_dir
mkdir $SLURM_TMPDIR/sub_${sub_num}_out
mkdir $SLURM_TMPDIR/image
mkdir $SLURM_TMPDIR/license
# Required fMRIprep files
cp ${project}/dataset_description.json $SLURM_TMPDIR/fmri_processing/results/TC2See
cp ${project}/fmriprep2.simg $SLURM_TMPDIR/image
cp ${project}/license.txt $SLURM_TMPDIR/license
apptainer run --cleanenv \
-B $SLURM_TMPDIR/fmri_processing/results/TC2See:/raw \
-B $SLURM_TMPDIR/sub_${sub_num}_out:/output \
-B $SLURM_TMPDIR/work_dir:/work_dir \
-B $SLURM_TMPDIR/image:/image \
-B $SLURM_TMPDIR/license:/license \
$SLURM_TMPDIR/image/fmriprep.simg \
/raw /output participant \
--participant-label ${sub_num} \
--work-dir /work_dir \
--fs-license-file /license/license.txt \
--output-spaces fsaverage \
--stop-on-first-crash
Version:
fMRIPrep 23.2.1
Environment (Docker, Singularity / Apptainer, custom installation):
I am using an image downloaded using:
singularity build fmriprep-23.2.1.simg docker://poldracklab/fmriprep:23.2.1
and this is being ran on an HPC cluster.
Data formatted according to a validatable standard? Please provide the output of the validator:
Relevant log outputs (up to 20 lines):
Node res_tmpl failed to run on host cdr482.int.cedar.computecanada.ca.
240313-23:45:36,666 nipype.workflow ERROR:
Saving crash info to /output/sub-10/log/20240313-234447_6c8dff82-48bf-45de-a65b-4731b65aaf7e/crash-20240313-234536-<my_username>-res_tmpl-6e55d8d7-b76e-4ad6-b989-591474ca4258.txt
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node res_tmpl.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/niworkflows/interfaces/nibabel.py", line 314, in _run_interface
resample_by_spacing(
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/niworkflows/utils/images.py", line 255, in resample_by_spacing
data = gaussian_filter(in_file.get_fdata(), smooth)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nibabel/dataobj_images.py", line 373, in get_fdata
data = np.asanyarray(self._dataobj, dtype=dtype)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nibabel/arrayproxy.py", line 439, in __array__
arr = self._get_scaled(dtype=dtype, slicer=())
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nibabel/arrayproxy.py", line 406, in _get_scaled
scaled = apply_read_scaling(self._get_unscaled(slicer=slicer), scl_slope, scl_inter)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nibabel/arrayproxy.py", line 376, in _get_unscaled
return array_from_file(
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nibabel/volumeutils.py", line 472, in array_from_file
raise OSError(
OSError: Expected 64364544 bytes, got 14000589 bytes from object
- could the file be damaged?
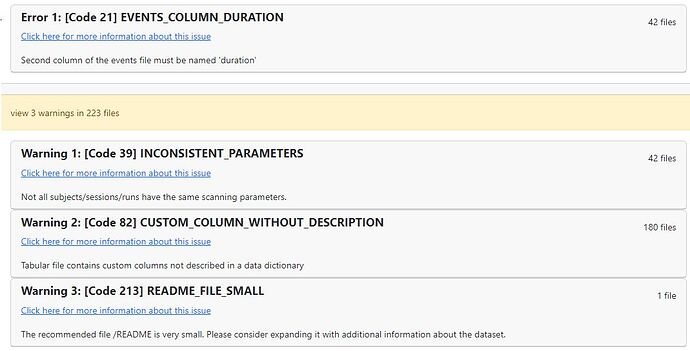
Screenshots / relevant information:
–cpus-per-task=16
–mem-per-cpu=16G
I have tried using up to 64G per cpu just in case but still had the same error.