Hi fMRIprep users,
I am trying to preprocess the openly available Balloon Analog Risk-Taking Task dataset available from OpenNeuro.
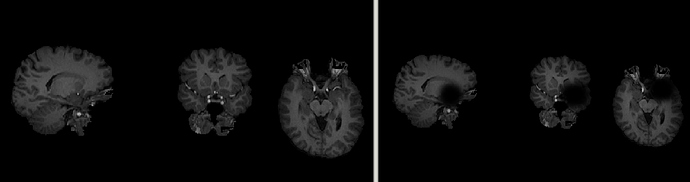
I have found that for one of the subjects, sub-04, preprocessing is failing. It seems as if the N4BiasFieldCorrection applied to the anatomical T1 image as part of the pipeline is actually inducing dropout at the front of the brain:
Left is the raw sub-04_T1w.nii.gz image, right is the sub-04_desc-preproc_T1w.nii.gz. As far as I’m aware the only difference between these images is that ANT’s N4BiasFieldCorrection has been applied to the right. This is subsequently causing skull-stripping to fail (brain mask it too small), and then registration to fail. Preprocessing is successful for all other subjects in the dataset.
I was wondering if it is possible to remove the N4BiasFieldCorrection from the pipeline, or if any other solutions exist? I have tried using the --force-bbr argument as was suggested here but the problem persisted.
I am running fMRIprep v1.5.1through singularity, here is the complete command:
/usr/local/miniconda/bin/fmriprep /well/nichols/users/bas627/BIDS_Data/RESULTS/SC2/data/raw/ds001_R2.0.4 /well/nichols/users/bas627/BIDS_Data/RESULTS/SC2/data/processed/ds001 participant --participant-label 04 --ignore slicetiming --fs-license-file /well/nichols/users/bas627/BIDS_Data/RESULTS/SC2/license.txt --fs-no-reconall -w /well/nichols/users/bas627/BIDS_Data/RESULTS/SC2/data/processed/ds001/ds001_sub-04_work --mem-mb 13000 --low-mem --resource-monitor --nthreads 4 -vvv
I am happy to provide more info if necessary. Thank you!
Alex