We hope to apply fMRIPrep on the QTAB dataset
QTAB includes MP2RAGE files with 2 different inversion times and 1 UNIT1. The author of this database applied 3dMPRAGEise. on these MP2RAGE files to denoise them and created *UNIT1_unbiased_clean.nii.gz in the derivative folder.
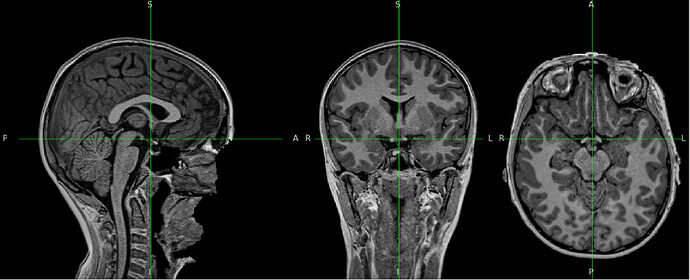
As an example, here is sub-0001_ses-01_UNIT1_unbiased_clean.nii.gz, which looks perfectly fine
fMRIPrep on sub-0001 ran successfully without any error, but the results are strange for t1w. We hope to hear your thoughts on this.
Let me explain what we did on the dataset and showed the fMRIPrep results below
For sub-0001, we then copied the
derivatives/UNIT1_denoised/sub-0001/ses-01/anat/sub-0001_ses-01_UNIT1_unbiased_clean.nii.gz
to
sub-0001//ses-01/anat/
(and also did the same thing for ses-02).
To create a JSON for this ‘t1w’ file, we then copied
sub-0001//ses-01/anat/sub-0001_ses-01_UNIT1_denoised.json
and changed its name to
sub-0001_ses-01_t1w.json.
(Note the UNIT1_denoised images are scans denoised straight from the scanner, but are generally still too noisy)
We just changed two things on its json sidecar:
from
"ImageComments": "DENOISED_IMAGE_(lambda_=_1)",
"SeriesDescription": "MP2RAGE_wip900C_VE11C_UNI-DEN",
to
"ImageComments": "3dMPRAGEise",
"SeriesDescription": "MP2RAGE_wip900C_VE11C_UNI-DEN_3dMPRAGEise",
Probably unrelated but we also changed a few things to the fmap, so that fMRIPrep can correct func images with fmap. They used pepolar fmap here.
For resting state, we followed this suggestion by copying bold files to the fmap folders:
sub-0001//ses-01/fmap/sub-0001_ses-01_task-rest_dir-AP_bold.nii.gz
and
sub-0001//ses-01/fmap/sub-0001_ses-01_task-rest_dir-PA_bold.nii.gz
We then set the
sub-0001_ses-01_task-rest_dir-AP_bold.json to be intended for PA_bold ,i.e., adding
"IntendedFor":"ses-01/fmap/sub-0001_ses-01_task-rest_dir-PA_bold.nii.gz",
"B0FieldIdentifier": "pepolar_fmap0"
and
sub-0001_ses-01_task-rest_dir-PA_bold.json to be intended for
AP_bold, i.e., adding
"IntendedFor":"ses-01/fmap/sub-0001_ses-01_task-rest_dir-AP_bold.nii.gz",
"B0FieldIdentifier": "pepolar_fmap0".
Below are their json files (Note that we didn’t change anything on the task fmri fmap files. From what we gather, fmriprep should be able to deal with this pepolar fmap.)
Also below are the file structure we have for this subject before running fmriprep version: 23.2.2. (Note we tried with or without ignoring t2w, and the results were similar).
/opt/conda/envs/fmriprep/bin/fmriprep /data /out participant --participant_label 0001 --write-graph --notrack --skip_bids_validation --work-dir /home/william/working_dir --ignore=slicetiming --ignore=t2w --cifti-output --fs-subjects-dir freesurfer
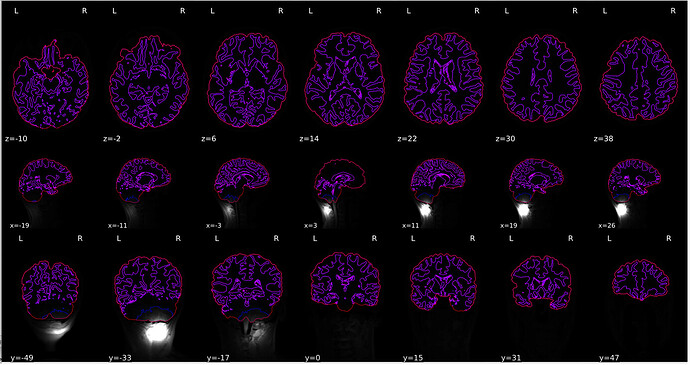
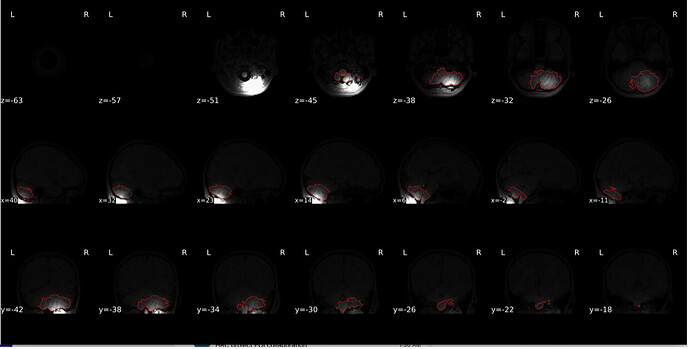
“Brain mask and brain tissue segmentation of the T1w” looks quite strange.
“Spatial normalization of the anatomical T1w reference” looks strange as well
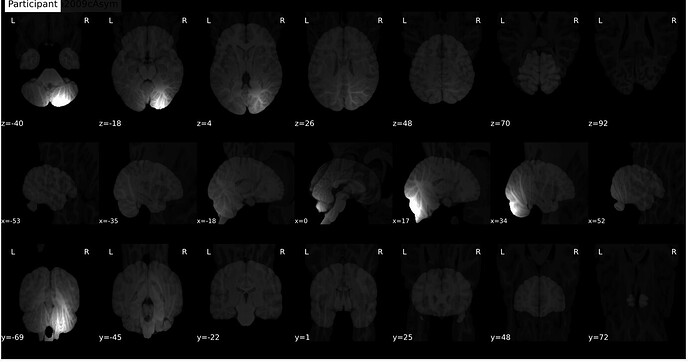
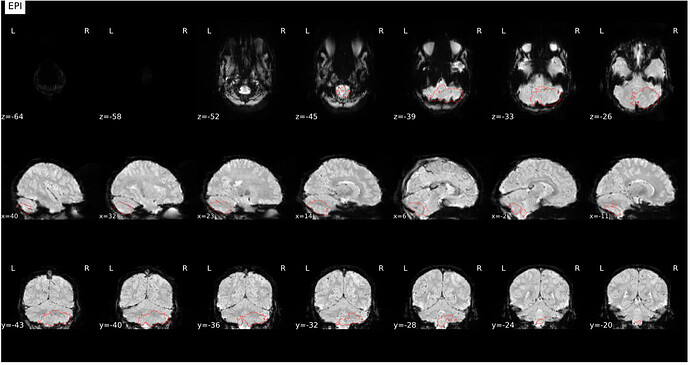
Surface reconstruction looks fine
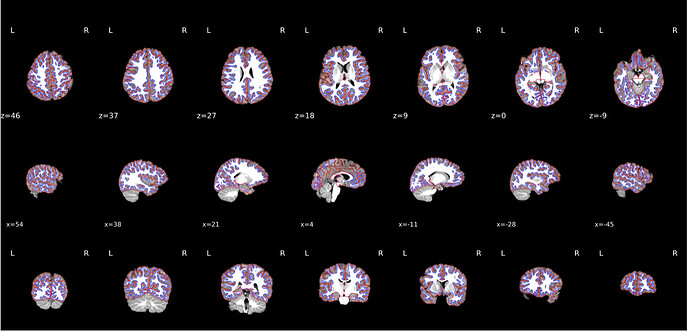
Alignment of functional and anatomical MRI data (coregistration) looks bad.
Maybe we missed something here?
Files:
sub-0001/
sub-0001//ses-01
sub-0001//ses-01/swi
sub-0001//ses-01/swi/sub-0001_ses-01_swi.json
sub-0001//ses-01/swi/sub-0001_ses-01_minIP.nii.gz
sub-0001//ses-01/swi/sub-0001_ses-01_part-phase_GRE.json
sub-0001//ses-01/swi/sub-0001_ses-01_part-mag_GRE.nii.gz
sub-0001//ses-01/swi/sub-0001_ses-01_swi.nii.gz
sub-0001//ses-01/swi/sub-0001_ses-01_part-phase_GRE.nii.gz
sub-0001//ses-01/swi/sub-0001_ses-01_part-mag_GRE.json
sub-0001//ses-01/swi/sub-0001_ses-01_minIP.json
sub-0001//ses-01/fmap
sub-0001//ses-01/fmap/sub-0001_ses-01_task-rest_dir-AP_bold.nii.gz
sub-0001//ses-01/fmap/sub-0001_ses-01_task-rest_dir-PA_bold.json
sub-0001//ses-01/fmap/sub-0001_ses-01_task-rest_dir-PA_bold.nii.gz
sub-0001//ses-01/fmap/sub-0001_ses-01_task-rest_dir-AP_bold.json
sub-0001//ses-01/func
sub-0001//ses-01/func/sub-0001_ses-01_task-rest_dir-AP_bold.nii.gz
sub-0001//ses-01/func/sub-0001_ses-01_task-rest_dir-PA_bold.json
sub-0001//ses-01/func/sub-0001_ses-01_task-rest_dir-PA_bold.nii.gz
sub-0001//ses-01/func/sub-0001_ses-01_task-rest_dir-AP_bold.json
sub-0001//ses-01/dwi
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-01_dwi.json
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-02_dwi.json
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-01_dwi.json
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-01_dwi.nii.gz
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-01_dwi.bvec
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-02_dwi.nii.gz
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-01_dwi.bval
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-01_dwi.nii.gz
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-02_dwi.bval
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-02_dwi.json
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-01_dwi.bval
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-02_dwi.nii.gz
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-01_dwi.bvec
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-02_dwi.bvec
sub-0001//ses-01/dwi/.goutputstream-7NL2M2
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-AP_run-02_dwi.bval
sub-0001//ses-01/dwi/sub-0001_ses-01_dir-PA_run-02_dwi.bvec
sub-0001//ses-01/anat
sub-0001//ses-01/anat/sub-0001_ses-01_FLAIR.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_UNIT1.json
sub-0001//ses-01/anat/sub-0001_ses-01_T2w_TSE_run-03.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_FLAIR.json
sub-0001//ses-01/anat/sub-0001_ses-01_UNIT1.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_UNIT1_denoised.json
sub-0001//ses-01/anat/sub-0001_ses-01_T2w.json
sub-0001//ses-01/anat/sub-0001_ses-01_inv-2_MP2RAGE_defacemask.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_T2w_TSE_run-02.json
sub-0001//ses-01/anat/sub-0001_ses-01_T2w_TSE_run-01.json
sub-0001//ses-01/anat/sub-0001_ses-01_inv-1_MP2RAGE.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_T2w.nii.gz
sub-0001//ses-01/anat/.goutputstream-HBPGN2
sub-0001//ses-01/anat/sub-0001_ses-01_UNIT1_denoised.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_T2w_TSE_run-03.json
sub-0001//ses-01/anat/sub-0001_ses-01_inv-1_MP2RAGE.json
sub-0001//ses-01/anat/sub-0001_ses-01_T2w_TSE_run-02.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_T2w_TSE_run-01.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_inv-2_MP2RAGE.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_inv-2_MP2RAGE.json
sub-0001//ses-01/anat/sub-0001_ses-01_T1w.nii.gz
sub-0001//ses-01/anat/sub-0001_ses-01_T1w.json
sub-0001//ses-01/perf
sub-0001//ses-01/perf/sub-0001_ses-01_asl.json
sub-0001//ses-01/perf/sub-0001_ses-01_m0scan.json
sub-0001//ses-01/perf/sub-0001_ses-01_m0scan.nii.gz
sub-0001//ses-01/perf/sub-0001_ses-01_aslcontext.tsv
sub-0001//ses-01/perf/sub-0001_ses-01_asl.nii.gz
sub-0001//ses-02
sub-0001//ses-02/swi
sub-0001//ses-02/swi/sub-0001_ses-02_part-phase_GRE.json
sub-0001//ses-02/swi/sub-0001_ses-02_part-phase_GRE.nii.gz
sub-0001//ses-02/swi/sub-0001_ses-02_part-mag_GRE.json
sub-0001//ses-02/swi/sub-0001_ses-02_swi.json
sub-0001//ses-02/swi/sub-0001_ses-02_minIP.json
sub-0001//ses-02/swi/sub-0001_ses-02_part-mag_GRE.nii.gz
sub-0001//ses-02/swi/sub-0001_ses-02_swi.nii.gz
sub-0001//ses-02/swi/sub-0001_ses-02_minIP.nii.gz
sub-0001//ses-02/fmap
sub-0001//ses-02/fmap/sub-0001_ses-02_task-rest_dir-PA_bold.json
sub-0001//ses-02/fmap/sub-0001_ses-02_dir-PA_epi.json
sub-0001//ses-02/fmap/sub-0001_ses-02_dir-PA_epi.nii.gz
sub-0001//ses-02/fmap/sub-0001_ses-02_task-rest_dir-AP_bold.nii.gz
sub-0001//ses-02/fmap/sub-0001_ses-02_task-rest_dir-AP_bold.json
sub-0001//ses-02/fmap/sub-0001_ses-02_dir-AP_epi.nii.gz
sub-0001//ses-02/fmap/sub-0001_ses-02_dir-AP_epi.json
sub-0001//ses-02/fmap/sub-0001_ses-02_task-rest_dir-PA_bold.nii.gz
sub-0001//ses-02/func
sub-0001//ses-02/func/sub-0001_ses-02_task-rest_dir-PA_bold.json
sub-0001//ses-02/func/sub-0001_ses-02_task-partlycloudy_events.json
sub-0001//ses-02/func/sub-0001_ses-02_task-rest_dir-AP_bold.nii.gz
sub-0001//ses-02/func/sub-0001_ses-02_task-partlycloudy_events.tsv
sub-0001//ses-02/func/sub-0001_ses-02_task-rest_dir-AP_bold.json
sub-0001//ses-02/func/sub-0001_ses-02_task-partlycloudy_bold.json
sub-0001//ses-02/func/sub-0001_ses-02_task-partlycloudy_bold.nii.gz
sub-0001//ses-02/func/sub-0001_ses-02_task-rest_dir-PA_bold.nii.gz
sub-0001//ses-02/dwi
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-01_dwi.bvec
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-02_dwi.bvec
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-02_dwi.bval
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-01_dwi.json
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-01_dwi.bval
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-02_dwi.bval
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-02_dwi.bvec
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-01_dwi.nii.gz
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-01_dwi.json
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-01_dwi.nii.gz
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-02_dwi.nii.gz
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-PA_run-02_dwi.json
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-02_dwi.nii.gz
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-01_dwi.bvec
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-02_dwi.json
sub-0001//ses-02/dwi/sub-0001_ses-02_dir-AP_run-01_dwi.bval
sub-0001//ses-02/anat
sub-0001//ses-02/anat/sub-0001_ses-02_T2w_TSE_run-02.json
sub-0001//ses-02/anat/sub-0001_ses-02_FLAIR.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_inv-2_MP2RAGE_defacemask.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_T2w_TSE_run-01.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_inv-1_MP2RAGE.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_UNIT1.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_T1w.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_inv-2_MP2RAGE.json
sub-0001//ses-02/anat/sub-0001_ses-02_inv-1_MP2RAGE.json
sub-0001//ses-02/anat/sub-0001_ses-02_T2w_TSE_run-01.json
sub-0001//ses-02/anat/sub-0001_ses-02_inv-2_MP2RAGE.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_UNIT1_denoised.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_T2w.json
sub-0001//ses-02/anat/sub-0001_ses-02_T1w.json
sub-0001//ses-02/anat/sub-0001_ses-02_UNIT1.json
sub-0001//ses-02/anat/sub-0001_ses-02_UNIT1_denoised.json
sub-0001//ses-02/anat/sub-0001_ses-02_FLAIR.json
sub-0001//ses-02/anat/sub-0001_ses-02_T2w_TSE_run-02.nii.gz
sub-0001//ses-02/anat/sub-0001_ses-02_T2w.nii.gz
sub-0001//ses-02/perf
sub-0001//ses-02/perf/sub-0001_ses-02_asl.json
sub-0001//ses-02/perf/sub-0001_ses-02_asl.nii.gz
sub-0001//ses-02/perf/sub-0001_ses-02_m0scan.json
sub-0001//ses-02/perf/sub-0001_ses-02_m0scan.nii.gz
sub-0001//ses-02/perf/sub-0001_ses-02_aslcontext.tsv
ses-01/anat/sub-0001_ses-01_T1w.json
{
"AcquisitionMatrixPE": 300,
"AcquisitionNumber": 1,
"AcquisitionTime": "11:20:19.000000",
"BaseResolution": 320,
"BodyPartExamined": "BRAIN",
"CoilString": "HC1-7;NC1_2",
"ConsistencyInfo": "N4_VE11C_LATEST_20160120",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20190410 GCC6.3.0",
"DeviceSerialNumber": "167001",
"DwellTime": 6.5e-06,
"EchoTime": 0.00299,
"ImageComments": "3dMPRAGEise",
"ImageOrientationPatientDICOM": [-0.0652318, 0.99787, 9.3308e-09, 0.0330828, 0.00216267, -0.99945],
"ImageType": [
"DERIVED",
"PRIMARY",
"M",
"UNI",
"DIS3D",
"DIS2D"
],
"ImagingFrequency": 123.229,
"InPlanePhaseEncodingDirectionDICOM": "ROW",
"InstitutionAddress": "Cooper_60_St._Lucia_QLD_AU_4072",
"InstitutionName": "St_Lucia_Campus",
"InstitutionalDepartmentName": "Gehrmann_Labs_MRI",
"MRAcquisitionType": "3D",
"MagneticFieldStrength": 3,
"Manufacturer": "Siemens",
"ManufacturersModelName": "Prisma_fit",
"Modality": "MR",
"PartialFourier": 0.75,
"PatientPosition": "HFS",
"PercentPhaseFOV": 93.75,
"PhaseEncodingSteps": 226,
"PhaseResolution": 1,
"PixelBandwidth": 240,
"ProcedureStepDescription": "Research_16092_QTAB",
"ProtocolName": "MP2RAGE_wip900C_VE11C",
"PulseSequenceDetails": "%CustomerSeq%_tfl_wip900C_VE11C",
"RawImage": false,
"ReceiveCoilActiveElements": "HC1-7;NC1,2",
"ReceiveCoilName": "HeadNeck_64",
"ReconMatrixPE": 300,
"RefLinesPE": 24,
"RepetitionTime": 4,
"SAR": 0.0695932,
"ScanOptions": "IR_PFP",
"ScanningSequence": "GR_IR",
"SequenceName": "tfl3d1_16ns",
"SequenceVariant": "SK_SP_MP",
"SeriesDescription": "MP2RAGE_wip900C_VE11C_UNI-DEN_3dMPRAGEise",
"SeriesNumber": 8,
"ShimSetting": [3036, -4179, -850, 100, -62, -225, 52, 12],
"SliceThickness": 0.8,
"SoftwareVersions": "syngo_MR_E11",
"StationName": "mrc_Trio",
"TxRefAmp": 206.955,
"WipMemBlock": "WIP_Identifier: WIP#900C"
}
ses-01/fmap/sub-0001_ses-01_task-rest_dir-AP_bold.json
{
"AcquisitionMatrixPE": 104,
"AcquisitionNumber": 1,
"AcquisitionTime": "11:28:16.240000",
"BandwidthPerPixelPhaseEncode": 17.17,
"BaseResolution": 104,
"BodyPartExamined": "BRAIN",
"ConsistencyInfo": "N4_VE11C_LATEST_20160120",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20171215 GCC6.3.0",
"DerivedVendorReportedEchoSpacing": 0.000560011,
"DeviceSerialNumber": "167001",
"DwellTime": 2.1e-06,
"EchoTime": 0.03,
"EchoTrainLength": 91,
"EffectiveEchoSpacing": 0.000560011,
"FlipAngle": 52,
"ImageOrientationPatientDICOM": [0.997551, 0.0664439, 0.0218547, -0.0550732, 0.938738, -0.340204],
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"ND",
"MOSAIC"
],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"InstitutionAddress": "Cooper_60_St._Lucia_QLD_AU_4072",
"InstitutionName": "St_Lucia_Campus",
"InstitutionalDepartmentName": "Gehrmann_Labs_MRI",
"MRAcquisitionType": "2D",
"MagneticFieldStrength": 3,
"Manufacturer": "Siemens",
"ManufacturersModelName": "Prisma_fit",
"Modality": "MR",
"MultibandAccelerationFactor": 6,
"PartialFourier": 0.875,
"PatientPosition": "HFS",
"PercentPhaseFOV": 100,
"PhaseEncodingDirection": "j-",
"PhaseEncodingSteps": 91,
"PhaseResolution": 1,
"PixelBandwidth": 2290,
"ProcedureStepDescription": "Research_16092_QTAB",
"ProtocolName": "ep2d_REST_SMS6_A-P",
"PulseSequenceDetails": "%SiemensSeq%_ep2d_bold",
"ReceiveCoilActiveElements": "HC1-7;NC1,2",
"ReceiveCoilName": "HeadNeck_64",
"ReconMatrixPE": 104,
"RepetitionTime": 0.93,
"SAR": 0.0683576,
"ScanOptions": "PFP_FS",
"ScanningSequence": "EP",
"SequenceName": "_epfid2d1_104",
"SequenceVariant": "SK",
"SeriesDescription": "ep2d_REST_SMS6_A-P",
"SeriesNumber": 9,
"ShimSetting": [3037, -4137, -746, 577, -94, 157, -69, 54],
"SliceThickness": 2,
"SliceTiming": [0.6825, 0, 0.455, 0.075, 0.53, 0.1525, 0.6075, 0.3025, 0.7575, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.075, 0.53, 0.1525, 0.6075, 0.3025, 0.7575, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.075, 0.53, 0.1525, 0.6075, 0.3025, 0.7575, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.075, 0.53, 0.1525, 0.6075, 0.3025, 0.7575, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.075, 0.53, 0.1525, 0.6075, 0.3025, 0.7575, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.075, 0.53, 0.1525, 0.6075, 0.3025, 0.7575, 0.38, 0.835, 0.2275],
"SoftwareVersions": "syngo_MR_E11",
"SpacingBetweenSlices": 2,
"StationName": "mrc_Trio",
"TaskName": "rest",
"TotalReadoutTime": 0.0576811,
"TxRefAmp": 206.955,
"IntendedFor":"ses-01/fmap/sub-0001_ses-01_task-rest_dir-PA_bold.nii.gz",
"B0FieldIdentifier": "pepolar_fmap0"
}
ses-01/fmap/sub-0001_ses-01_task-rest_dir-PA_bold.json
{
"AcquisitionMatrixPE": 104,
"AcquisitionNumber": 1,
"AcquisitionTime": "11:33:56.935000",
"BandwidthPerPixelPhaseEncode": 17.17,
"BaseResolution": 104,
"BodyPartExamined": "BRAIN",
"ConsistencyInfo": "N4_VE11C_LATEST_20160120",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20171215 GCC6.3.0",
"DerivedVendorReportedEchoSpacing": 0.000560011,
"DeviceSerialNumber": "167001",
"DwellTime": 2.1e-06,
"EchoTime": 0.03,
"EchoTrainLength": 91,
"EffectiveEchoSpacing": 0.000560011,
"FlipAngle": 52,
"ImageOrientationPatientDICOM": [0.997551, 0.0664439, 0.0218547, -0.0550732, 0.938738, -0.340204],
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"ND",
"MOSAIC"
],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"InstitutionAddress": "Cooper_60_St._Lucia_QLD_AU_4072",
"InstitutionName": "St_Lucia_Campus",
"InstitutionalDepartmentName": "Gehrmann_Labs_MRI",
"MRAcquisitionType": "2D",
"MagneticFieldStrength": 3,
"Manufacturer": "Siemens",
"ManufacturersModelName": "Prisma_fit",
"Modality": "MR",
"MultibandAccelerationFactor": 6,
"PartialFourier": 0.875,
"PatientPosition": "HFS",
"PercentPhaseFOV": 100,
"PhaseEncodingDirection": "j",
"PhaseEncodingSteps": 91,
"PhaseResolution": 1,
"PixelBandwidth": 2290,
"ProcedureStepDescription": "Research_16092_QTAB",
"ProtocolName": "ep2d_REST_SMS6_P-A",
"PulseSequenceDetails": "%SiemensSeq%_ep2d_bold",
"ReceiveCoilActiveElements": "HC1-7;NC1,2",
"ReceiveCoilName": "HeadNeck_64",
"ReconMatrixPE": 104,
"RepetitionTime": 0.93,
"SAR": 0.0683576,
"ScanOptions": "PFP_FS",
"ScanningSequence": "EP",
"SequenceName": "_epfid2d1_104",
"SequenceVariant": "SK",
"SeriesDescription": "ep2d_REST_SMS6_P-A",
"SeriesNumber": 10,
"ShimSetting": [3037, -4137, -746, 577, -94, 157, -69, 54],
"SliceThickness": 2,
"SliceTiming": [0.6825, 0, 0.455, 0.0775, 0.5325, 0.1525, 0.6075, 0.305, 0.76, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.0775, 0.5325, 0.1525, 0.6075, 0.305, 0.76, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.0775, 0.5325, 0.1525, 0.6075, 0.305, 0.76, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.0775, 0.5325, 0.1525, 0.6075, 0.305, 0.76, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.0775, 0.5325, 0.1525, 0.6075, 0.305, 0.76, 0.38, 0.835, 0.2275, 0.6825, 0, 0.455, 0.0775, 0.5325, 0.1525, 0.6075, 0.305, 0.76, 0.38, 0.835, 0.2275],
"SoftwareVersions": "syngo_MR_E11",
"SpacingBetweenSlices": 2,
"StationName": "mrc_Trio",
"TaskName": "rest",
"TotalReadoutTime": 0.0576811,
"TxRefAmp": 206.955,
"IntendedFor":"ses-01/fmap/sub-0001_ses-01_task-rest_dir-AP_bold.nii.gz",
"B0FieldIdentifier": "pepolar_fmap0"
}
@tsalo I tagged you here since I saw your post on QTAB. I would hope to hear your thoughts on this.
Thanks so much.
Narun