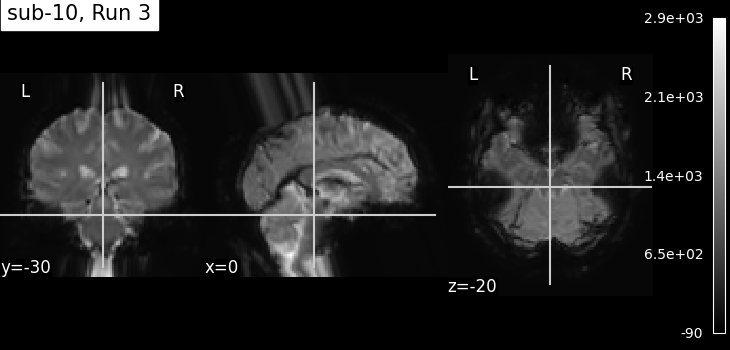
Summary of what happened:
Hi all,
I have some artifacts generated after preprocessing in fmriprep. It seems like there is something going wrong in the preprocessing that pulls/stretches the brain down from the brainstem and up from the motor cortex above it. Please see the image of preprocessed data.
In the html reports, everything seems normal though.
Currently only the default paramteres are used (fMRIPrep version: 25.1.3) and this seems to be prevalent among the whole data set. Any idea as to whty this is happening?
I am analysing fmri data for the first time and any help is much appreciated!
Thanks a lot in advance,
Ann
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep-docker $bids_root_dir $bids_root_dir/derivatives \
participant \
--participant-label $subj \
--md-only-boilerplate \
--ignore fieldmaps \
--fs-license-file $FS_LICENSE\
--fs-no-reconall \
--output-spaces MNI152NLin2009cAsym:res-2 \
--nthreads $nthreads \
--stop-on-first-crash \
--mem_mb $mem_mb \
--skip-bids-validation
Version:
25.1.3
Environment (Docker, Singularity / Apptainer, custom installation):
DOCKER with fmriprep-docker
Data formatted according to a validatable standard? Please provide the output of the validator:
not yet checked
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE