Summary of what happened:
Hey everyone,
I am processing a 3D EPI with a limited field of view (FoV; acquired on a Siemens scanner with TR of 0.9s) with fmriprep 25.1.3. The processing works without errors however, when looking at the processed functional image (sub_ses_task-task_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii), I can see that the signal has been stretched outside of the field of view (FoV).
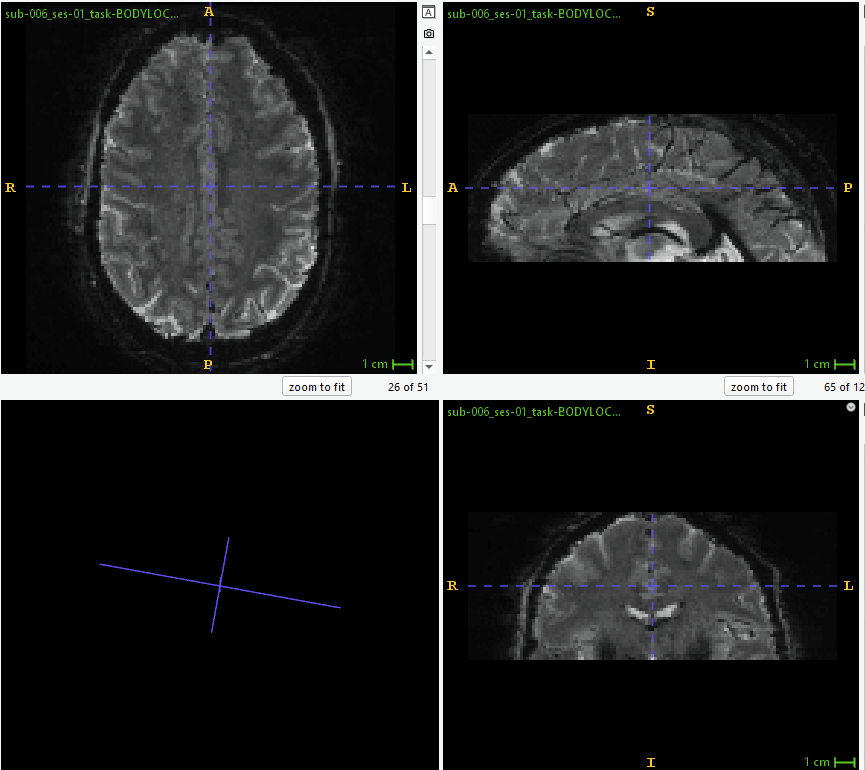
See below for a screenshot from sub_ses_task-task_run-1_desc-coreg_boldref.nii (for reference, how the EPI looks without signal stretched outside of FoV; using ITK Snap 4.2.2):
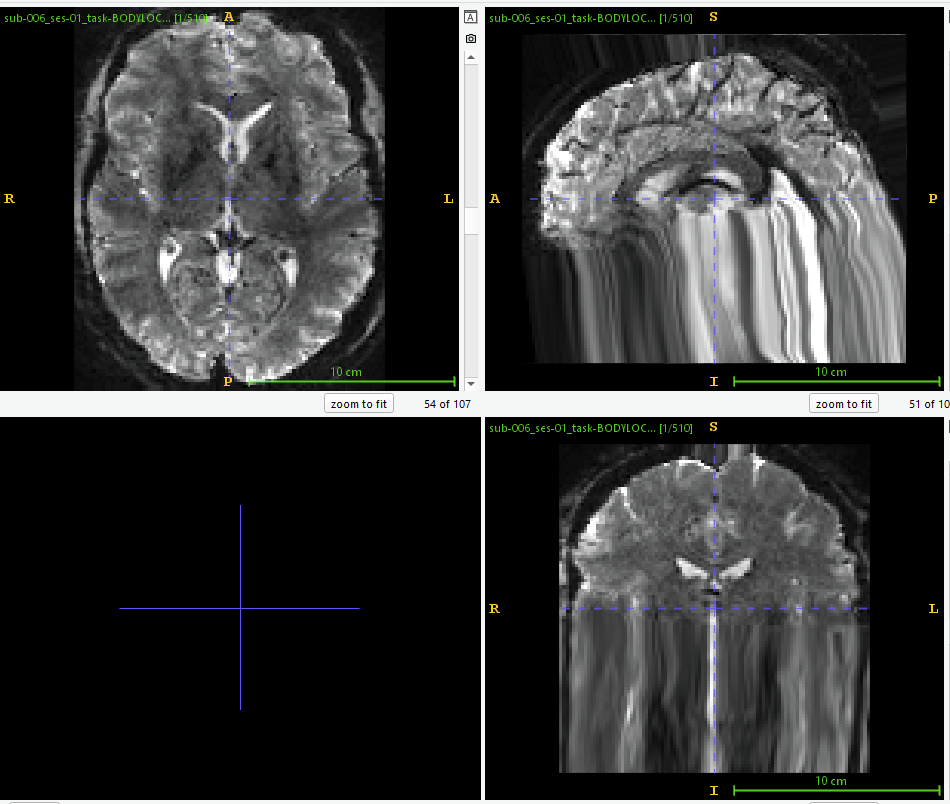
and a screenshot from ‘sub_ses_task-task_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii’ (using ITK Snap 4.2.2):
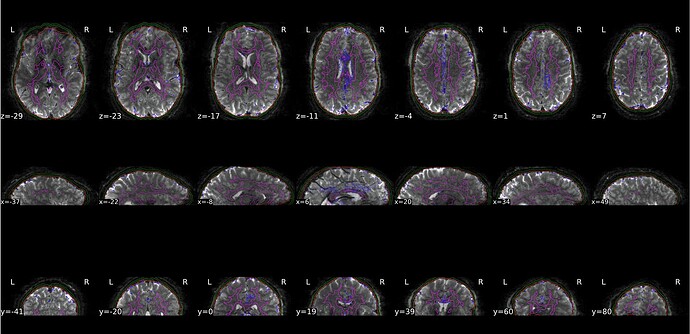
Here is a screenshot from the HTML report (‘Brain mask and (anatomical/temporal) CompCor ROIs’ section) where there is no stretching outside of the FoV of the EPI:
I do get an EPI mask after processing that I can apply. However, I am concerned that the bottom layers are stretched to fill the space outside of the FoV, so after applying the mask, the volume would still contain some of the stretched signal and I would lose signal that is being stretched outside of the FoV.
I am wondering what is going wrong and how I can fix it.
Any help would be massively appreciated!
Command used (and if a helper script was used, a link to the helper script or the command generated):
Here is the fmriprep command:
/opt/conda/envs/fmriprep/bin/fmriprep \
--skip_bids_validation \
--participant-label 006 \
--nprocs 25 \
--mem 30720 \
--ignore fieldmaps slicetiming \
--fs-license-file /data/Code/fmriprep/license.txt \
--fs-subjects-dir /data/Code/fmriprep/tmp-freesurfer \
-w /data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01/wrk-dir \
--write-graph \
--notrack \
--aggregate-session-reports 1 \
/data/Data/data/mri/bids_bodymap \
/data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01 \
participant
The Freesurfer folder referenced in the code has been made by a previous fmriprep run on the same subject.
Version:
25.1.3
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
Here are the contents of the log:
[environment]
cpu_count = 28
exec_env = "docker"
free_mem = 26.2
overcommit_policy = "always"
overcommit_limit = "50%"
nipype_version = "1.10.0"
templateflow_version = "24.2.2"
version = "25.1.3"
[execution]
bids_dir = "/data/Data/data/mri/bids_bodymap"
bids_database_dir = "/data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01/wrk-dir/20250923-033007_fd9bc862-83fb-4f9a-9625-58562aa2728e/bids_db"
bids_description_hash = "43c0ca6052b6d6ce24770ea6d38016a9c98b41bee2f4d128b58e3f5b0d97370d"
boilerplate_only = false
sloppy = false
debug = []
fmriprep_dir = "/data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01"
fs_license_file = "/data/Code/fmriprep/license.txt"
fs_subjects_dir = "/data/Code/fmriprep/tmp-freesurfer"
layout = "BIDS Layout: .../data/mri/bids_bodymap | Subjects: 1 | Sessions: 1 | Runs: 1"
log_dir = "/data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01/logs"
log_level = 25
low_mem = false
md_only_boilerplate = false
notrack = true
track_carbon = false
country_code = "CAN"
output_dir = "/data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01"
me_output_echos = false
aggr_ses_reports = 1
output_layout = "bids"
output_spaces = "MNI152NLin2009cAsym:res-native"
reports_only = false
run_uuid = "20250923-033007_fd9bc862-83fb-4f9a-9625-58562aa2728e"
participant_label = [ "006",]
templateflow_home = "/home/jovyan/.cache/templateflow"
work_dir = "/data/Code/fmriprep/script-test/min-example-v6/sub-006/ses-01/wrk-dir"
write_graph = true
[workflow]
anat_only = false
bold2anat_dof = 6
bold2anat_init = "t1w"
cifti_output = false
fmap_bspline = false
force_syn = false
hires = true
fs_no_resume = false
ignore = [ "fieldmaps", "slicetiming",]
force = []
level = "full"
longitudinal = false
run_msmsulc = true
medial_surface_nan = false
project_goodvoxels = false
regressors_all_comps = false
regressors_dvars_th = 1.5
regressors_fd_th = 0.5
run_reconall = true
skull_strip_fixed_seed = false
skull_strip_template = "OASIS30ANTs"
skull_strip_t1w = "force"
slice_time_ref = 0.5
spaces = "MNI152NLin2009cAsym:res-native"
use_syn_sdc = false
me_t2s_fit_method = "curvefit"
[nipype]
crashfile_format = "txt"
get_linked_libs = false
memory_gb = 30.72
nprocs = 25
omp_nthreads = 8
plugin = "MultiProc"
remove_unnecessary_outputs = true
resource_monitor = false
stop_on_first_crash = false
[seeds]
master = 62265
ants = 7591
numpy = 16589
[execution.derivatives]
[execution.dataset_links]
raw = "/data/Data/data/mri/bids_bodymap"
templateflow = "/home/jovyan/.cache/templateflow"
[nipype.plugin_args]
maxtasksperchild = 1
raise_insufficient = false
Screenshots / relevant information:
I tried running fmriprep with these additional options without luck.
1) --use-syn-sdc warn --force syn-sdc
2) --force no-bbr --project-goodvoxels --use-syn-sdc warn --force syn-sdc
3) --force bbr --project-goodvoxels --use-syn-sdc warn --force syn-sdc
4) --force bbr --project-goodvoxels --skull-strip-fixed-seed --random-seed 12345 --use-syn-sdc warn --force syn-sdc