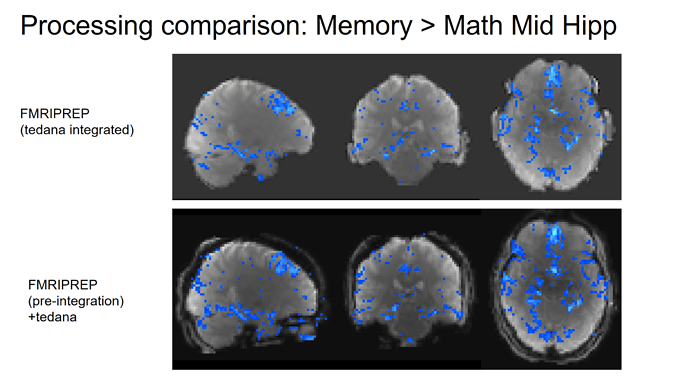
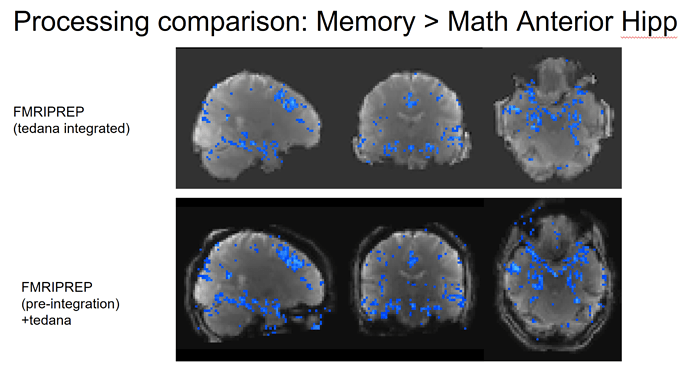
Hi NeuroStars,
New to both fmriprep and tedana (& multiecho in general). We’ve been able to successfully run fmriprep on some multiecho data on our cluster (yay!) but are getting the error below when we try to use tedana. It’s not immediately clear to us what this error means (e.g. is it an issue with the data? the preproc? the inputs?). See call below the error. We’ve also tried running without masks and with multiple masks, but don’t feel familiar enough with the other flags to go with anything other than the defaults. Any thoughts/advice greatly appreciated!
[failed] tedana output
attempting to run tedana for sub-06ME
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use arr[tuple(seq)] instead of arr[seq]. In the future this will be interpreted as an array index, arr[np.array(seq)], which will result either in an error or a different result.
return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/model/fit.py:161: RuntimeWarning: invalid value encountered in true_divide
F_S0 = (alpha - SSE_S0) * (n_echos - 1) / (SSE_S0)
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/model/fit.py:168: RuntimeWarning: invalid value encountered in true_divide
F_R2 = (alpha - SSE_R2) * (n_echos - 1) / (SSE_R2)
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/model/fit.py:177: RuntimeWarning: invalid value encountered in greater
F_S0[F_S0 > F_MAX] = F_MAX
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/model/fit.py:178: RuntimeWarning: invalid value encountered in greater
F_R2[F_R2 > F_MAX] = F_MAX
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/model/fit.py:161: RuntimeWarning: divide by zero encountered in true_divide
F_S0 = (alpha - SSE_S0) * (n_echos - 1) / (SSE_S0)
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/sklearn/cross_validation.py:41: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. Also note that the interface of the new CV iterators are different from that of this module. This module will be removed in 0.20.
“This module will be removed in 0.20.”, DeprecationWarning)
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/sklearn/ensemble/weight_boosting.py:29: DeprecationWarning: numpy.core.umath_tests is an internal NumPy module and should not be imported. It will be removed in a future NumPy release.
from numpy.core.umath_tests import inner1d
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/sklearn/grid_search.py:42: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. This module will be removed in 0.20.
DeprecationWarning)
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/sklearn/learning_curve.py:22: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the functions are moved. This module will be removed in 0.20
DeprecationWarning)
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/scipy/stats/stats.py:2253: RuntimeWarning: invalid value encountered in true_divide
return (a - mns) / sstd
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use arr[tuple(seq)] instead of arr[seq]. In the future this will be interpreted as an array index, arr[np.array(seq)], which will result either in an error or a different result.
return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/selection/select_comps.py:555: RuntimeWarning: invalid value encountered in greater
np.union1d(np.union1d(all_comps[spz > 1],
/share/apps/tedana/0.0.4/lib/python3.6/site-packages/tedana/selection/select_comps.py:628: RuntimeWarning: invalid value encountered in greater
np.union1d(np.union1d(all_comps[spz > 1],
Extremely limited reliable BOLD signal space! Not filtering components beyond BOLD/non-BOLD guesses.
No BOLD components detected! Please check data and results!
tedana call
Echo1={WorkDir}/{fmriprepdir}/fmriprep/{SUBJ_ID}/func/sub-06ME_task-mid_acq-normal_rec-magnitude_run-01_echo-1_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
Echo2={WorkDir}/{fmriprepdir}/fmriprep/{SUBJ_ID}/func/sub-06ME_task-mid_acq-normal_rec-magnitude_run-01_echo-2_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
Echo3={WorkDir}/{fmriprepdir}/fmriprep/{SUBJ_ID}/func/sub-06ME_task-mid_acq-normal_rec-magnitude_run-01_echo-3_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
Echo4={WorkDir}/{fmriprepdir}/fmriprep/{SUBJ_ID}/func/sub-06ME_task-mid_acq-normal_rec-magnitude_run-01_echo-4_bold_space-MNI152NLin2009cAsym_preproc.nii.gz
Mask1=${WorkDir}/${fmriprepdir}/fmriprep/${SUBJ_ID}/func/sub-06ME_task-mem_acq-normal_rec-magnitude_run-01_echo-1_bold_space-MNI152NLin2009cAsym_brainmask.nii.gz
tedana -d ${Echo1} ${Echo2} ${Echo3} ${Echo4} -e 12.2 29.48 46.76 64.04 --mask ${Mask1} --label ${SUBJ_ID}_tedana
 so the two mid runs seem fine, but the two mem runs crash with the following error:
so the two mid runs seem fine, but the two mem runs crash with the following error: