Hi everyone,
I am working with high-resolution rs-fmri data. I am trying to register the bold images to MNI using FSL by
1.registering bold to T1 using FLIRT
2.T1 to MNI with FLIRT(dof 9)
3.T1 to MNI with FNIRT using the transformation from the 2nd step
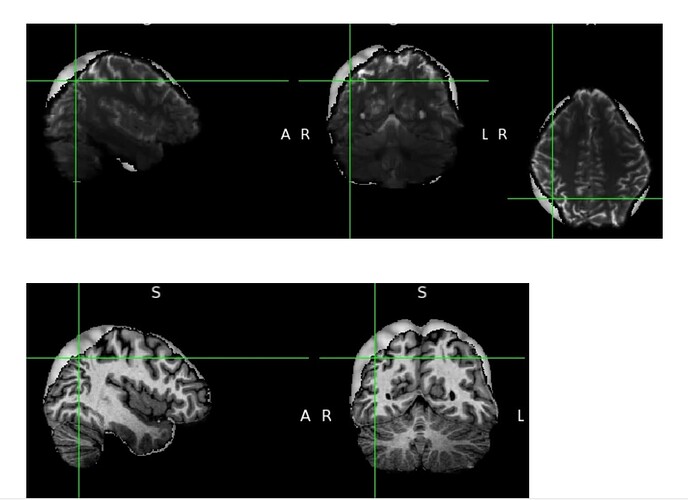
4. Applying to warp from the 3rd to the bold images, along with the transformation matrix from the 1st step. However, I get weird results specifically around the lateral top parts of the cortex, near(ish) to the boundaries, both with the T1 and the bold (see the screenshots below, both overlaid on the MNI 1mm template).Medial and inferior looks fine (not shown)
I tried resolving the issue by changing some of the parameters/options and arguments as below. As a reference, I used the default MNI_2mm template configuration file in FSL. However, this did not make much difference. I am aware this configuration file is for 2mm space, however I do not know which parameters would optimize registration into the 1mm space. Does anyone have an idea or have used FNIRT to normalize into 1mm space?
I am using fsl/6.0.5.1. Voxel sizes; bold 1.6 mm iso, and T1 0.73mm iso.
Any help would be appreciated! Thank you!
fnirt --ref=${FSLDIR}/data/standard/MNI152_T1_1mm.nii.gz
–in=sub-01_T1w.nii.gz
–aff=T1_to_ref_aff.mat
–cout=warp_T1_2_mni.nii.gz
–refmask=MNI152_T1_1mm_brain_mask_dil.nii.gz
–inmask=sub-01-brain_mask.nii.gz --imprefm=1 --impinm=1
–imprefval=0 --impinval=0 -
-subsamp=4,4,2,2,1,1 --miter=5,5,5,5,5,10
–infwhm=8,6,5,4.5,3,2 --reffwhm=8,6,5,4,2,0
–lambda=300,150,100,50,40,30 --estint=1,1,1,1,1,0
–applyrefmask=0,0,0,0,1,1 --applyinmask=1
–warpres=10,10,10 --ssqlambda=1 --regmod=bending_energy
–intmod=global_non_linear_with_bias --intorder=5 --biasres=50,50,50
–biaslambda=10000 --refderiv=0