Dear experts,
I am running FEAT (in FSL) which includes unwarping of the functional images with a Siemens GRE fieldmap. I have prepared the fieldmap images as instructed in the documentation (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FUGUE/Guide#SIEMENS_data).
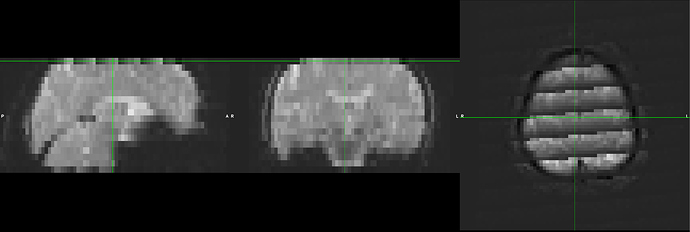
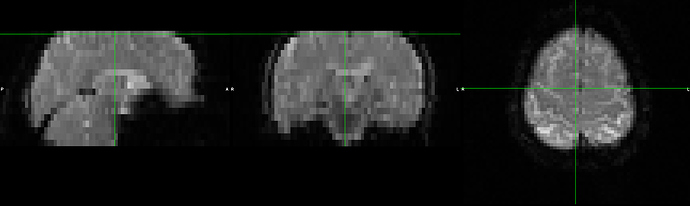
After examination of the resultant FEAT output and temporary files, I noticed that the unwarping and co-registration steps to the anatomical images are introducing “grid-like” artifacts to the top and bottom slices of the functional runs, and is consistent across subjects. Inspection of the report logs (and with fsleyes) reveal it has corrected (to some degree) the susceptibility inhomogeneities. Here is the example_func (undistorted) image:
And the example example_func_distorted:
As you will probably note this is an older acquisition where the voxel resolution in the sagittal plane (6mm) is larger than typical sequences (multiband acceleration was not available here), leading to incomplete brain coverage across most subjects.
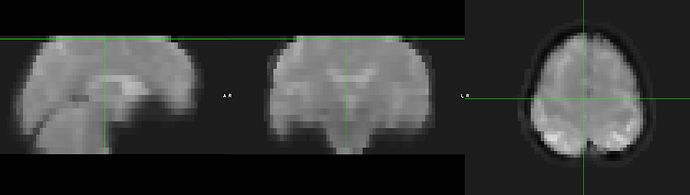
The issue appears to occur between calling of the epi_reg and applywarp functions within FEAT preprocessing stage 1, where the example_func_undistorted image is created. Contrary to some older posts (https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=fsl;fc64216.1411), the applywarp commands proceeding straight after mcflirt also did not introduce these artifacts into the final filtered_func images etc. For example, see:
Given that the prefiltered_func_data and resultant filtered_func_data do not contain these slice artifiacts, I am wondering if later analysis steps using this data are going to be contaminated?
We’ve run ICA (melodic) to see if these noise components will be present, and noted the spatial and temporal characteristics of the components are similar across images with and without distortion correction.
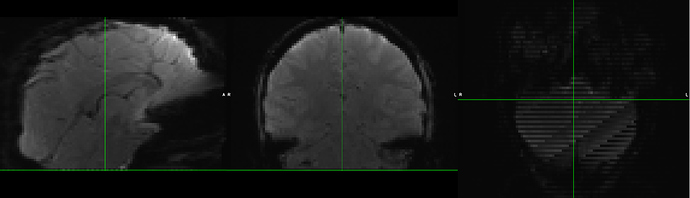
The larger voxel resolution and incomplete brain coverage will evidently lead to these rather noisy voxels at the top and bottom slices. We would prefer not to remove such voxels from further analysis, for obvious reasons. We have nonetheless run the same FEAT parameters on high-resolution data (from the same scanner), and are still experiencing this gridding at the top/bottom (particularly) slices - but anyway for some reason the distortion correction hasn’t worked that well:
All the available logs are found here: