Dear esperts,
I’m having trouble processing multicenter fMRI data. I used the methods of ComBat to analyze the differences in ReHo and fALFF values between MDD and healthy controls.
My data is a small sample with three different sites in total. I first started with a pre-process and generated 90% groupmasks by EPI for 68,400 voxels, and obtained 68,400 (ReHo or fALFF values) x n matrix as raw data. Then I put subjects’ age, sex, head movement, and type of disease (“MDD” and “HCs”) as covariates. Finally, I convert the matrix after ComBat to nii. and conduct statistical analysis.
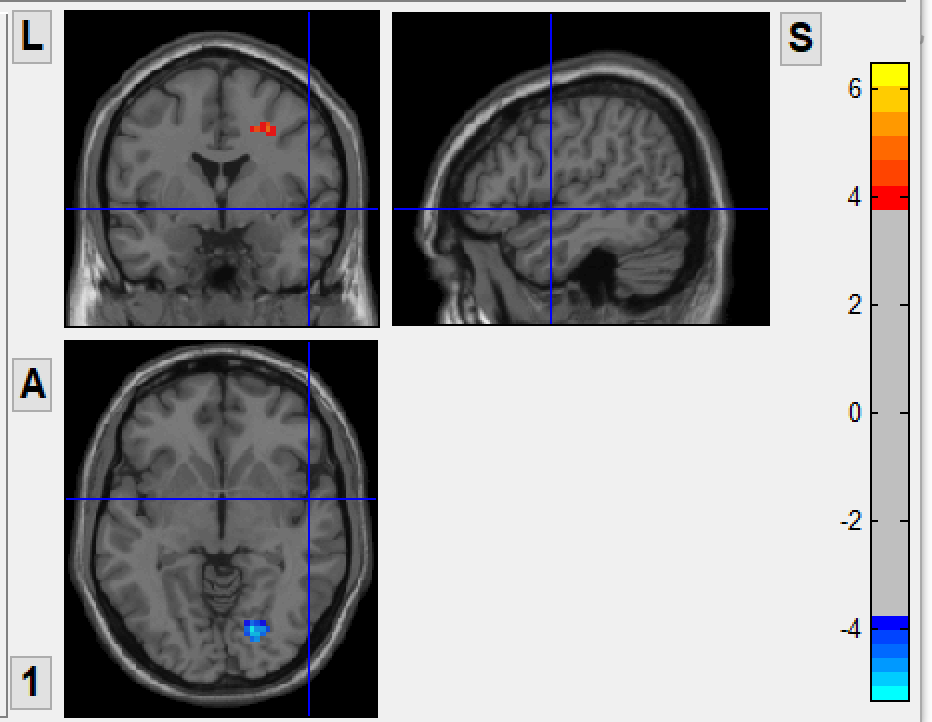
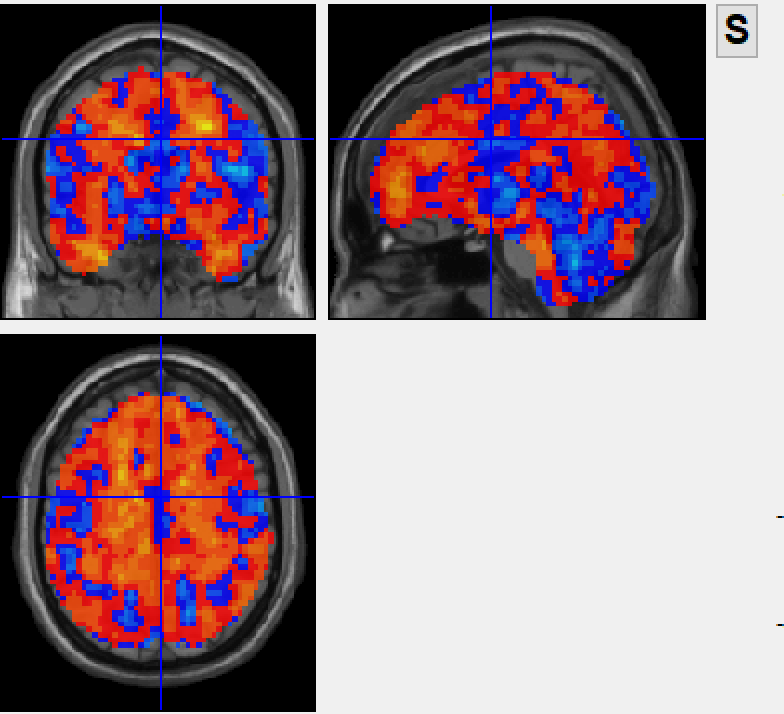
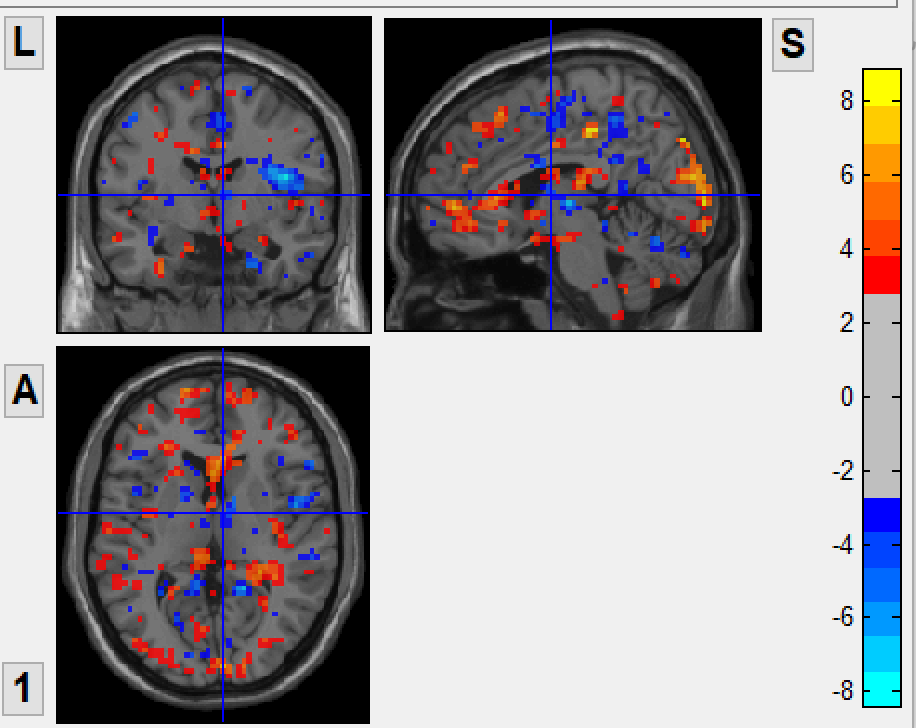
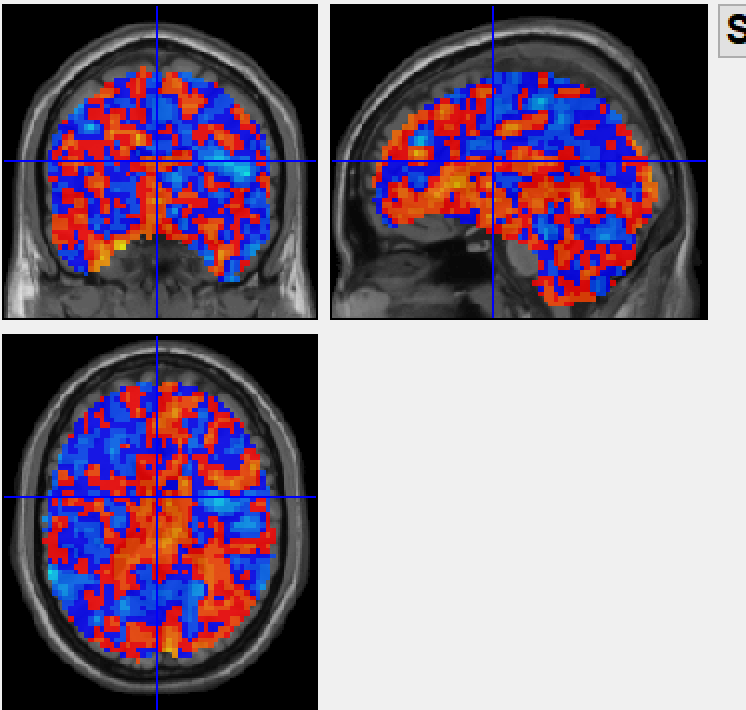
However, I found significant differences in the whole brain using ComBat including brainstem and ventricles after FDR correction. t-value maps were not as continuous as they were before combat processing, but were in small clumps.
Is anyone else had the same problem with this, or could anyone provide me a better solution?
My handling and code is mainly based on this article:10.1016/j.neuroimage.2023.120089
Best,
Fangzhou Guo
before combat
after combat