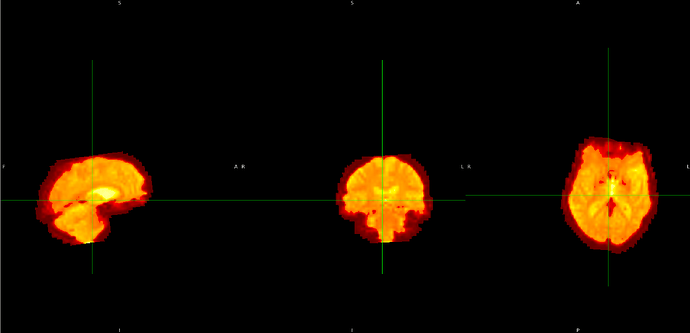
Summary of what happened:
I am currently working on preprocessing images that will be used in a model using FSL. However, I noticed that some parts of the fmri_in_mni image appear slightly cropped. After reviewing the intermediate images, I found that the issue starts from the flirt_fmri_to_t1 image, where a small part of the image seems to be cut off. Could you please advise on how to handle this situation? Any guidance would be greatly appreciated.
Command used (and if a helper script was used, a link to the helper script or the command generated):
import os
import subprocess
import nibabel as nib
import time
# Environment setup
os.environ['FSLDIR'] = '/path/to/fsl'
os.environ['PATH'] += os.pathsep + os.path.join(os.environ['FSLDIR'], 'bin')
# File paths (redacted)
fmri_file = '/path/to/fmri_data.nii.gz'
t1_file = '/path/to/t1_image.nii.gz'
mni_template = os.path.join(os.environ['FSLDIR'], 'data', 'standard', 'MNI152_T1_1mm_brain.nii.gz')
output_dir = '/path/to/output_directory'
os.makedirs(output_dir, exist_ok=True)
def run_command(command):
"""Utility function to run shell commands"""
print(f"Running: {command}")
subprocess.run(command, shell=True, check=True)
# 1. Load the resting-state fMRI file
print("Loading fMRI data...")
fmri_img = nib.load(fmri_file)
# 2. BET: Perform brain extraction on fMRI data
print("Starting BET brain extraction for fMRI...")
bet_fmri_output = os.path.join(output_dir, "fmri_brain")
bet_command = f"bet {fmri_file} {bet_fmri_output} -F -f 0.5 -m"
run_command(bet_command)
# 2.1 Extract the first volume from the fMRI data
print("Extracting the first volume from fMRI data...")
fmri_img = nib.load(bet_fmri_output + ".nii.gz")
first_volume_data = fmri_img.get_fdata()[..., 0] # Extract the first volume
first_volume_img = nib.Nifti1Image(first_volume_data, fmri_img.affine, fmri_img.header)
first_volume_path = os.path.join(output_dir, "fmri_brain_first_volume.nii.gz")
nib.save(first_volume_img, first_volume_path)
# 3. BET: Perform brain extraction on the T1-weighted image
print("Starting BET brain extraction for T1w...")
bet_t1_output = os.path.join(output_dir, "t1_brain")
bet_t1_command = f"bet {t1_file} {bet_t1_output} -f 0.5 -m"
run_command(bet_t1_command)
# 4. FLIRT: Perform linear registration of the brain-extracted fMRI data to the brain-extracted T1w image
print("Starting FLIRT registration (fMRI -> T1w)...")
flirt_fmri_to_t1_output = os.path.join(output_dir, "flirt_fmri_to_t1")
flirt_fmri_to_t1_mat_output = os.path.join(output_dir, "fmri_to_t1.mat")
flirt_fmri_to_t1_command = f"flirt -in {first_volume_path} -ref {bet_t1_output} -out {flirt_fmri_to_t1_output} -omat {flirt_fmri_to_t1_mat_output} -dof 6"
run_command(flirt_fmri_to_t1_command)
# 5. FLIRT: Perform linear registration of the brain-extracted T1w image to the MNI152 template
print("Starting FLIRT registration (T1w -> MNI)...")
flirt_t1_to_mni_output = os.path.join(output_dir, "flirt_t1_to_mni")
flirt_t1_to_mni_mat_output = os.path.join(output_dir, "t1_to_mni.mat")
flirt_t1_to_mni_command = f"flirt -in {bet_t1_output} -ref {mni_template} -out {flirt_t1_to_mni_output} -omat {flirt_t1_to_mni_mat_output} -dof 12"
run_command(flirt_t1_to_mni_command)
# 6. FNIRT: Perform non-linear registration of the T1w image to the MNI152 template
print("Starting FNIRT registration (T1w -> MNI)...")
fnirt_output = os.path.join(output_dir, "t1_to_mni_warp")
fnirt_command = f"fnirt --in={bet_t1_output} --ref={mni_template} --aff={flirt_t1_to_mni_mat_output} --cout={fnirt_output}"
run_command(fnirt_command)
# 7. applywarp: Apply the warp from FNIRT to transform the fMRI data into MNI152 template space
print("Applying warp to fMRI data to MNI space...")
apply_warp_output = os.path.join(output_dir, "fmri_in_mni")
apply_warp_command = f"applywarp --in={flirt_fmri_to_t1_output} --ref={mni_template} --warp={fnirt_output} --out={apply_warp_output}"
run_command(apply_warp_command)
print("fMRI preprocessing pipeline completed successfully.")
Screenshots / relevant information:
- flirt_fmri_to_t1