Hi everyone,
I used AFNI following the tutorial from the link below, Correlate Brain and Behavior in AFNI, and used the 3dTcorr1D function to obtain the corr_visual file. When I view the results in the AFNI GUI, how should I set the threshold? For example, should the p-value be set to 0.05? Also, how should I set the cluster size?
https://blog.cogneurostats.com/2013/03/08/correlating-brain-and-behavior-in-afni/
Thank you for your help!
Best regards,
Shuning
Hi, Shuning-
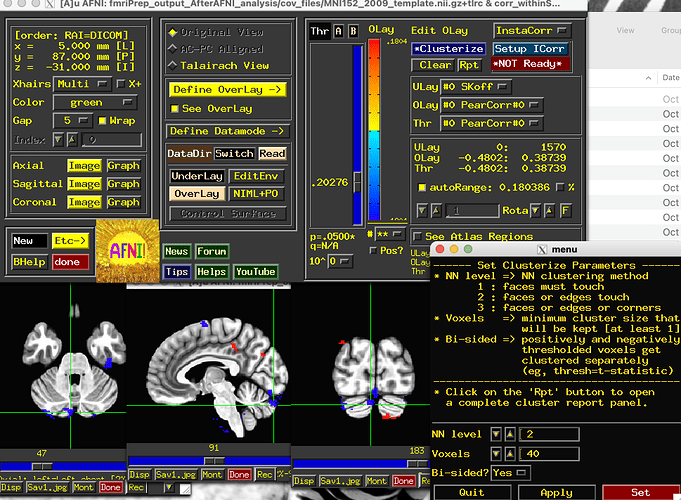
Clustering is a large topic. Some aspects of it are discussed here. In general, it involves setting 2 thresholds: a voxelwise threshold and a cluster size threshold.
In AFNI, the basic way to estimate the latter according to assumptions of approximate noise smoothness is:
- run “3dFWHMx …” with
-ACF .. and -mask .. on the residuals of your data, to estimate the spatial extent of approximate+average noise smoothness across the brain;
- then run 3dClustSim using that same mask and also using the ACF parameters output by 3dFWHMx, to do cluster simulations to estimate a lookup table for cluster size threshold for a given voxelwise threshold (in terms of p-value) and familywise error rate (FWE, commonly called the alpha value) that you want to control at.
But importantly, this is a very rough approximation for second-level (cluster) correction. Thresholding in general can hide a lot of useful things, and so using transparent thresholding is important for more informative results reporting.
–pt