Dear all,
I am new to heudicov, I followed the tutorial by Stanford Center for Reproducible Neuroscience (http://reproducibility.stanford.edu/bids-tutorial-series-part-2a/) to convert my own dataset. It seems that there is no error, but obviously one resting scan was not converted.
Information of my system: ubuntu 18.04 (dual boot with window 10), wiht the latest docker version of heudiconv.
Appreciate any comments and suggestions, because I read the tutorial carefully and could’t find which part was wrong
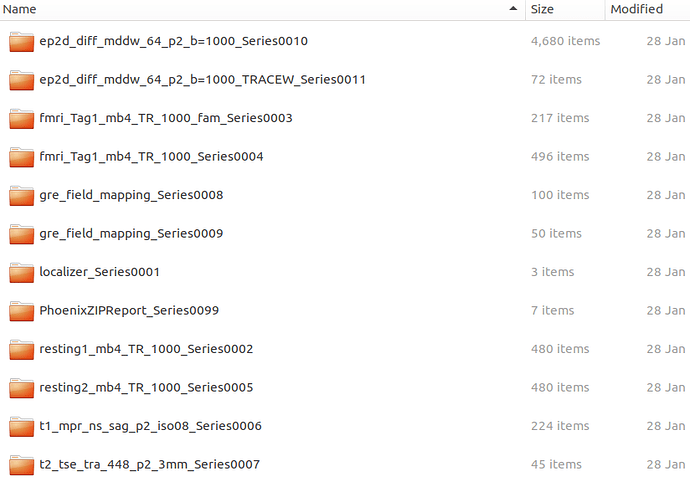
Below is part of dicominfor.tsv ('cause I can only upload one image)
| dim1 | dim2 | dim3 | dim4 | TR | TE | protocol_name |
|---|---|---|---|---|---|---|
| 512 | 512 | 3 | 1 | 0.0086 | 4 | localizer |
| 72 | 84 | 60 | 480 | 1 | 29 | resting1_mb4_TR_1000 |
| 72 | 84 | 60 | 217 | 1 | 29 | fmri_Tag1_mb4_TR_1000_fam |
| 72 | 84 | 60 | 496 | 1 | 29 | fmri_Tag1_mb4_TR_1000 |
| 72 | 84 | 60 | 480 | 1 | 29 | resting2_mb4_TR_1000 |
| 320 | 320 | 224 | 1 | 1.9 | 2.54 | t1_mpr_ns_sag_p2_iso08 |
| 448 | 448 | 45 | 1 | 6.1 | 79 | t2_tse_tra_448_p2_3mm |
| 64 | 64 | 100 | 1 | 1.03 | 7.35 | gre_field_mapping |
| 64 | 64 | 50 | 1 | 1.03 | 7.35 | gre_field_mapping |
| 128 | 128 | 4680 | 1 | 9.1 | 85 | ep2d_diff_mddw_64_p2_b=1000 |
| 128 | 128 | 72 | 1 | 9.1 | 85 | ep2d_diff_mddw_64_p2_b=1000 |
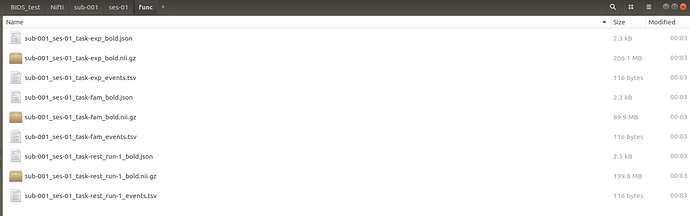
Below is the screen shot of the func folder, as you can see, only one run of resting there, but the dicom data have two runs.
Here is the code in my heuristic.py
import os
def create_key(template, outtype=('nii.gz',), annotation_classes=None):
if template is None or not template:
raise ValueError('Template must be a valid format string')
return template, outtype, annotation_classes
def infotodict(seqinfo):
"""Heuristic evaluator for determining which runs belong where
"""
t1w = create_key('sub-{subject}/{session}/anat/sub-{subject}_T1w')
t2w = create_key('sub-{subject}/{session}/anat/sub-{subject}_T2w')
func_fam = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-fam_bold')
func_task = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-exp_bold')
func_rest = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-{item:01d}_bold')
fmap_mag = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_magnitude')
fmap_phase = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_phasediff')
dwi = create_key('sub-{subject}/{session}/dwi/sub-{subject}_{session}_run-{item:01d}_dwi')
# data = create_key('run{item:03d}')
info = {t1w: [], t2w: [], func_fam: [], func_task: [], func_rest: [], fmap_mag: [], fmap_phase:[], dwi: []}
last_run = len(seqinfo)
for s in seqinfo:
"""
The namedtuple `s` contains the following fields:
"""
# define T1w
if (s.dim1 == 320) and (s.dim2 == 320) and ('t1_mpr' in s.protocol_name):
info[t1w] = [s.series_id]
# define T2w
if (s.dim1 == 448) and (s.dim2 == 448) and ('t2_tse' in s.protocol_name):
info[t2w] = [s.series_id]
# define functional familiarization
if (s.dim1 == 72) and (s.dim4 == 217) and ('fmri' in s.protocol_name):
info[func_fam] = [s.series_id]
# define functional task
if (s.dim1 == 72) and (s.dim4 == 496) and ('fmri' in s.protocol_name):
info[func_task] = [s.series_id]
# define functional resting
if (s.dim1 == 72) and (s.dim2 == 84) and ('resting' in s.protocol_name):
info[func_rest] = [s.series_id]
# define field map (magnitude)
if (s.dim3 == 100) and (s.dim4 == 1) and ('gre_field_mapping' in s.protocol_name):
info[fmap_mag] = [s.series_id]
# define field map (phasic)
if (s.dim3 == 50) and (s.dim4 == 1) and ('gre_field_mapping' in s.protocol_name):
info[fmap_phase] = [s.series_id]
# define DTI
if (s.dim1 == 128) and (s.dim4 == 1) and ('diff' in s.protocol_name):
info[dwi].append(s.series_id)
return info
Below are the code and output when I run heudiconv:
hcp4715@hcp4715-Precision-5510:~$ sudo docker run --rm -it -v /media/hcp4715/USB1/Data/RepDopa/BIDS_test:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}// -o /base/Nifti/ -f /base/Nifti/code/heuristic.py -s 001 -ss 01 -c dcm2niix -b --overwrite --minmeta
INFO: Running heudiconv version 0.5.4.dev1
INFO: Need to process 1 study sessions
INFO: PROCESSING STARTS: {‘subject’: ‘001’, ‘outdir’: ‘/base/Nifti/’, ‘session’: ‘01’}
INFO: Processing 6854 dicoms
INFO: Will not reuse existing conversion table files because heuristic has changed
INFO: Analyzing 6854 dicoms
INFO: Generated sequence info with 11 entries
INFO: Doing conversion using dcm2niix
INFO: Converting /base/Nifti/sub-001/ses-01/anat/sub-001_ses-01_T1w (224 DICOMs) -> /base/Nifti/sub-001/ses-01/anat . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:28,7 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niixjbel5e38/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niixjbel5e38/convert”.
190212-23:03:28,95 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f anat -o . -s n -v n /tmp/dcm2niixjbel5e38/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f anat -o . -s n -v n /tmp/dcm2niixjbel5e38/convert
190212-23:03:28,311 nipype.interface INFO:
stdout 2019-02-12T23:03:28.311461:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:28.311461:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:28,311 nipype.interface INFO:
stdout 2019-02-12T23:03:28.311461:Found 224 DICOM file(s)
INFO: stdout 2019-02-12T23:03:28.311461:Found 224 DICOM file(s)
190212-23:03:28,311 nipype.interface INFO:
stdout 2019-02-12T23:03:28.311461:Convert 224 DICOM as ./anat (320x320x224x1)
INFO: stdout 2019-02-12T23:03:28.311461:Convert 224 DICOM as ./anat (320x320x224x1)
190212-23:03:29,95 nipype.interface INFO:
stdout 2019-02-12T23:03:29.095953:compress: “/usr/bin/pigz” -n -f -6 “./anat.nii”
INFO: stdout 2019-02-12T23:03:29.095953:compress: “/usr/bin/pigz” -n -f -6 “./anat.nii”
190212-23:03:29,96 nipype.interface INFO:
stdout 2019-02-12T23:03:29.095953:Conversion required 0.955019 seconds (0.295855 for core code).
INFO: stdout 2019-02-12T23:03:29.095953:Conversion required 0.955019 seconds (0.295855 for core code).
190212-23:03:29,125 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:29,412 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmetaruuj143z/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmetaruuj143z/embedder”.
190212-23:03:29,450 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:29,456 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/anat/sub-001_ses-01_T1w.json file
INFO: Converting /base/Nifti/sub-001/ses-01/anat/sub-001_ses-01_T2w (45 DICOMs) -> /base/Nifti/sub-001/ses-01/anat . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:29,461 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niixz9rsujso/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niixz9rsujso/convert”.
190212-23:03:29,481 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f anat -o . -s n -v n /tmp/dcm2niixz9rsujso/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f anat -o . -s n -v n /tmp/dcm2niixz9rsujso/convert
190212-23:03:29,534 nipype.interface INFO:
stdout 2019-02-12T23:03:29.534329:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:29.534329:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:29,534 nipype.interface INFO:
stdout 2019-02-12T23:03:29.534329:Found 45 DICOM file(s)
INFO: stdout 2019-02-12T23:03:29.534329:Found 45 DICOM file(s)
190212-23:03:29,534 nipype.interface INFO:
stdout 2019-02-12T23:03:29.534329:Convert 45 DICOM as ./anat (448x448x45x1)
INFO: stdout 2019-02-12T23:03:29.534329:Convert 45 DICOM as ./anat (448x448x45x1)
190212-23:03:29,784 nipype.interface INFO:
stdout 2019-02-12T23:03:29.784202:compress: “/usr/bin/pigz” -n -f -6 “./anat.nii”
INFO: stdout 2019-02-12T23:03:29.784202:compress: “/usr/bin/pigz” -n -f -6 “./anat.nii”
190212-23:03:29,784 nipype.interface INFO:
stdout 2019-02-12T23:03:29.784202:Conversion required 0.292782 seconds (0.048126 for core code).
INFO: stdout 2019-02-12T23:03:29.784202:Conversion required 0.292782 seconds (0.048126 for core code).
190212-23:03:29,801 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:30,43 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmetasqe8g9j9/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmetasqe8g9j9/embedder”.
190212-23:03:30,53 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:30,57 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/anat/sub-001_ses-01_T2w.json file
INFO: Converting /base/Nifti/sub-001/ses-01/func/sub-001_ses-01_task-fam_bold (217 DICOMs) -> /base/Nifti/sub-001/ses-01/func . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:30,66 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niixwo8mij39/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niixwo8mij39/convert”.
190212-23:03:30,151 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f func -o . -s n -v n /tmp/dcm2niixwo8mij39/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f func -o . -s n -v n /tmp/dcm2niixwo8mij39/convert
190212-23:03:30,413 nipype.interface INFO:
stdout 2019-02-12T23:03:30.413450:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:30.413450:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:30,413 nipype.interface INFO:
stdout 2019-02-12T23:03:30.413450:Found 217 DICOM file(s)
INFO: stdout 2019-02-12T23:03:30.413450:Found 217 DICOM file(s)
190212-23:03:30,413 nipype.interface INFO:
stdout 2019-02-12T23:03:30.413450:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
INFO: stdout 2019-02-12T23:03:30.413450:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
190212-23:03:30,414 nipype.interface INFO:
stdout 2019-02-12T23:03:30.413450:Convert 217 DICOM as ./func (84x72x60x217)
INFO: stdout 2019-02-12T23:03:30.413450:Convert 217 DICOM as ./func (84x72x60x217)
190212-23:03:32,700 nipype.interface INFO:
stdout 2019-02-12T23:03:32.700203:compress: “/usr/bin/pigz” -n -f -6 “./func.nii”
INFO: stdout 2019-02-12T23:03:32.700203:compress: “/usr/bin/pigz” -n -f -6 “./func.nii”
190212-23:03:32,700 nipype.interface INFO:
stdout 2019-02-12T23:03:32.700203:Conversion required 2.539633 seconds (0.319509 for core code).
INFO: stdout 2019-02-12T23:03:32.700203:Conversion required 2.539633 seconds (0.319509 for core code).
190212-23:03:32,728 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:33,359 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmetah7drbdhx/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmetah7drbdhx/embedder”.
190212-23:03:33,391 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:33,397 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/func/sub-001_ses-01_task-fam_bold.json file
INFO: Converting /base/Nifti/sub-001/ses-01/func/sub-001_ses-01_task-exp_bold (496 DICOMs) -> /base/Nifti/sub-001/ses-01/func . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:33,415 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niixhsbjkjop/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niixhsbjkjop/convert”.
190212-23:03:33,599 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f func -o . -s n -v n /tmp/dcm2niixhsbjkjop/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f func -o . -s n -v n /tmp/dcm2niixhsbjkjop/convert
190212-23:03:34,169 nipype.interface INFO:
stdout 2019-02-12T23:03:34.168945:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:34.168945:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:34,169 nipype.interface INFO:
stdout 2019-02-12T23:03:34.168945:Found 496 DICOM file(s)
INFO: stdout 2019-02-12T23:03:34.168945:Found 496 DICOM file(s)
190212-23:03:34,169 nipype.interface INFO:
stdout 2019-02-12T23:03:34.168945:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
INFO: stdout 2019-02-12T23:03:34.168945:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
190212-23:03:34,169 nipype.interface INFO:
stdout 2019-02-12T23:03:34.168945:Convert 496 DICOM as ./func (84x72x60x496)
INFO: stdout 2019-02-12T23:03:34.168945:Convert 496 DICOM as ./func (84x72x60x496)
190212-23:03:39,405 nipype.interface INFO:
stdout 2019-02-12T23:03:39.405333:compress: “/usr/bin/pigz” -n -f -6 “./func.nii”
INFO: stdout 2019-02-12T23:03:39.405333:compress: “/usr/bin/pigz” -n -f -6 “./func.nii”
190212-23:03:39,405 nipype.interface INFO:
stdout 2019-02-12T23:03:39.405333:Conversion required 5.796813 seconds (0.702813 for core code).
INFO: stdout 2019-02-12T23:03:39.405333:Conversion required 5.796813 seconds (0.702813 for core code).
190212-23:03:39,448 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:40,471 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmeta2k8pykfj/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmeta2k8pykfj/embedder”.
190212-23:03:40,542 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:40,549 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/func/sub-001_ses-01_task-exp_bold.json file
INFO: Converting /base/Nifti/sub-001/ses-01/func/sub-001_ses-01_task-rest_run-1_bold (480 DICOMs) -> /base/Nifti/sub-001/ses-01/func . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:40,566 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niix7vml9cft/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niix7vml9cft/convert”.
190212-23:03:40,756 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f func -o . -s n -v n /tmp/dcm2niix7vml9cft/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f func -o . -s n -v n /tmp/dcm2niix7vml9cft/convert
190212-23:03:41,307 nipype.interface INFO:
stdout 2019-02-12T23:03:41.307827:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:41.307827:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:41,308 nipype.interface INFO:
stdout 2019-02-12T23:03:41.307827:Found 480 DICOM file(s)
INFO: stdout 2019-02-12T23:03:41.307827:Found 480 DICOM file(s)
190212-23:03:41,308 nipype.interface INFO:
stdout 2019-02-12T23:03:41.307827:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
INFO: stdout 2019-02-12T23:03:41.307827:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
190212-23:03:41,308 nipype.interface INFO:
stdout 2019-02-12T23:03:41.307827:Convert 480 DICOM as ./func (84x72x60x480)
INFO: stdout 2019-02-12T23:03:41.307827:Convert 480 DICOM as ./func (84x72x60x480)
190212-23:03:46,350 nipype.interface INFO:
stdout 2019-02-12T23:03:46.350402:compress: “/usr/bin/pigz” -n -f -6 “./func.nii”
INFO: stdout 2019-02-12T23:03:46.350402:compress: “/usr/bin/pigz” -n -f -6 “./func.nii”
190212-23:03:46,350 nipype.interface INFO:
stdout 2019-02-12T23:03:46.350402:Conversion required 5.584305 seconds (0.687461 for core code).
INFO: stdout 2019-02-12T23:03:46.350402:Conversion required 5.584305 seconds (0.687461 for core code).
190212-23:03:46,393 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:47,427 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmetaq4dbbpsm/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmetaq4dbbpsm/embedder”.
190212-23:03:47,496 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:47,504 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/func/sub-001_ses-01_task-rest_run-1_bold.json file
INFO: Converting /base/Nifti/sub-001/ses-01/fmap/sub-001_ses-01_magnitude (100 DICOMs) -> /base/Nifti/sub-001/ses-01/fmap . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:47,509 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niixphmybaqi/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niixphmybaqi/convert”.
190212-23:03:47,551 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f fmap -o . -s n -v n /tmp/dcm2niixphmybaqi/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f fmap -o . -s n -v n /tmp/dcm2niixphmybaqi/convert
190212-23:03:47,623 nipype.interface INFO:
stdout 2019-02-12T23:03:47.623814:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:47.623814:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:47,624 nipype.interface INFO:
stdout 2019-02-12T23:03:47.623814:Found 100 DICOM file(s)
INFO: stdout 2019-02-12T23:03:47.623814:Found 100 DICOM file(s)
190212-23:03:47,624 nipype.interface INFO:
stdout 2019-02-12T23:03:47.623814:slices not stacked: echo varies (TE 4.89, 7.35; echo 1, 2). Use ‘merge 2D slices’ option to force stacking
INFO: stdout 2019-02-12T23:03:47.623814:slices not stacked: echo varies (TE 4.89, 7.35; echo 1, 2). Use ‘merge 2D slices’ option to force stacking
190212-23:03:47,624 nipype.interface INFO:
stdout 2019-02-12T23:03:47.623814:Convert 50 DICOM as ./fmap_e1 (64x64x50x1)
INFO: stdout 2019-02-12T23:03:47.623814:Convert 50 DICOM as ./fmap_e1 (64x64x50x1)
190212-23:03:47,642 nipype.interface INFO:
stdout 2019-02-12T23:03:47.642738:compress: “/usr/bin/pigz” -n -f -6 “./fmap_e1.nii”
INFO: stdout 2019-02-12T23:03:47.642738:compress: “/usr/bin/pigz” -n -f -6 “./fmap_e1.nii”
190212-23:03:47,642 nipype.interface INFO:
stdout 2019-02-12T23:03:47.642738:slices not stacked: echo varies (TE 7.35, 4.89; echo 2, 1). Use ‘merge 2D slices’ option to force stacking
INFO: stdout 2019-02-12T23:03:47.642738:slices not stacked: echo varies (TE 7.35, 4.89; echo 2, 1). Use ‘merge 2D slices’ option to force stacking
190212-23:03:47,643 nipype.interface INFO:
stdout 2019-02-12T23:03:47.642738:Convert 50 DICOM as ./fmap_e2 (64x64x50x1)
INFO: stdout 2019-02-12T23:03:47.642738:Convert 50 DICOM as ./fmap_e2 (64x64x50x1)
190212-23:03:47,657 nipype.interface INFO:
stdout 2019-02-12T23:03:47.657945:compress: “/usr/bin/pigz” -n -f -6 “./fmap_e2.nii”
INFO: stdout 2019-02-12T23:03:47.657945:compress: “/usr/bin/pigz” -n -f -6 “./fmap_e2.nii”
190212-23:03:47,658 nipype.interface INFO:
stdout 2019-02-12T23:03:47.657945:Conversion required 0.096665 seconds (0.057536 for core code).
INFO: stdout 2019-02-12T23:03:47.657945:Conversion required 0.096665 seconds (0.057536 for core code).
190212-23:03:47,678 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
WARNING: For now not embedding BIDS and info generated .nii.gz itself since sequence produced multiple files
INFO: Converting /base/Nifti/sub-001/ses-01/fmap/sub-001_ses-01_phasediff (50 DICOMs) -> /base/Nifti/sub-001/ses-01/fmap . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:47,693 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niix9yp7yhh1/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niix9yp7yhh1/convert”.
190212-23:03:47,715 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f fmap -o . -s n -v n /tmp/dcm2niix9yp7yhh1/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f fmap -o . -s n -v n /tmp/dcm2niix9yp7yhh1/convert
190212-23:03:47,758 nipype.interface INFO:
stdout 2019-02-12T23:03:47.758376:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:47.758376:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:47,758 nipype.interface INFO:
stdout 2019-02-12T23:03:47.758376:Found 50 DICOM file(s)
INFO: stdout 2019-02-12T23:03:47.758376:Found 50 DICOM file(s)
190212-23:03:47,758 nipype.interface INFO:
stdout 2019-02-12T23:03:47.758376:Convert 50 DICOM as ./fmap_e2_ph (64x64x50x1)
INFO: stdout 2019-02-12T23:03:47.758376:Convert 50 DICOM as ./fmap_e2_ph (64x64x50x1)
190212-23:03:47,769 nipype.interface INFO:
stdout 2019-02-12T23:03:47.769150:compress: “/usr/bin/pigz” -n -f -6 “./fmap_e2_ph.nii”
INFO: stdout 2019-02-12T23:03:47.769150:compress: “/usr/bin/pigz” -n -f -6 “./fmap_e2_ph.nii”
190212-23:03:47,769 nipype.interface INFO:
stdout 2019-02-12T23:03:47.769150:Conversion required 0.044770 seconds (0.029837 for core code).
INFO: stdout 2019-02-12T23:03:47.769150:Conversion required 0.044770 seconds (0.029837 for core code).
190212-23:03:47,788 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:47,800 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmeta2hh4m1pd/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmeta2hh4m1pd/embedder”.
190212-23:03:47,810 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:47,814 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/fmap/sub-001_ses-01_phasediff.json file
INFO: Converting /base/Nifti/sub-001/ses-01/dwi/sub-001_ses-01_run-1_dwi (4680 DICOMs) -> /base/Nifti/sub-001/ses-01/dwi . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:47,975 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niix5il0o9uz/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niix5il0o9uz/convert”.
190212-23:03:49,826 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f dwi -o . -s n -v n /tmp/dcm2niix5il0o9uz/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f dwi -o . -s n -v n /tmp/dcm2niix5il0o9uz/convert
190212-23:03:52,844 nipype.interface INFO:
stdout 2019-02-12T23:03:52.844433:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:52.844433:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:52,844 nipype.interface INFO:
stdout 2019-02-12T23:03:52.844433:Found 4680 DICOM file(s)
INFO: stdout 2019-02-12T23:03:52.844433:Found 4680 DICOM file(s)
190212-23:03:52,845 nipype.interface INFO:
stdout 2019-02-12T23:03:52.844433:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
INFO: stdout 2019-02-12T23:03:52.844433:slices stacked despite varying acquisition numbers (if this is not desired please recompile)
190212-23:03:52,845 nipype.interface INFO:
stdout 2019-02-12T23:03:52.844433:Warning: Saving 65 DTI gradients. Validate vectors (image slice orientation not reported, e.g. 2001,100B).
INFO: stdout 2019-02-12T23:03:52.844433:Warning: Saving 65 DTI gradients. Validate vectors (image slice orientation not reported, e.g. 2001,100B).
190212-23:03:52,845 nipype.interface INFO:
stdout 2019-02-12T23:03:52.844433:Convert 4680 DICOM as ./dwi (128x128x72x65)
INFO: stdout 2019-02-12T23:03:52.844433:Convert 4680 DICOM as ./dwi (128x128x72x65)
190212-23:03:55,202 nipype.interface INFO:
stdout 2019-02-12T23:03:55.202795:compress: “/usr/bin/pigz” -n -f -6 “./dwi.nii”
INFO: stdout 2019-02-12T23:03:55.202795:compress: “/usr/bin/pigz” -n -f -6 “./dwi.nii”
190212-23:03:55,202 nipype.interface INFO:
stdout 2019-02-12T23:03:55.202795:Conversion required 5.358445 seconds (2.609329 for core code).
INFO: stdout 2019-02-12T23:03:55.202795:Conversion required 5.358445 seconds (2.609329 for core code).
190212-23:03:55,484 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:55,785 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmeta75hmjz7q/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmeta75hmjz7q/embedder”.
190212-23:03:56,368 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:56,406 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/dwi/sub-001_ses-01_run-1_dwi.json file
INFO: Converting /base/Nifti/sub-001/ses-01/dwi/sub-001_ses-01_run-2_dwi (72 DICOMs) -> /base/Nifti/sub-001/ses-01/dwi . Converter: dcm2niix . Output types: (‘nii.gz’,)
190212-23:03:56,411 nipype.workflow INFO:
[Node] Setting-up “convert” in “/tmp/dcm2niixmsc8j9o_/convert”.
INFO: [Node] Setting-up “convert” in “/tmp/dcm2niixmsc8j9o_/convert”.
190212-23:03:56,441 nipype.workflow INFO:
[Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f dwi -o . -s n -v n /tmp/dcm2niixmsc8j9o_/convert
INFO: [Node] Running “convert” (“nipype.interfaces.dcm2nii.Dcm2niix”), a CommandLine Interface with command:
dcm2niix -b y -z y -x n -t n -m n -f dwi -o . -s n -v n /tmp/dcm2niixmsc8j9o_/convert
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
INFO: stdout 2019-02-12T23:03:56.504069:Chris Rorden’s dcm2niiX version v1.0.20180622 GCC6.3.0 (64-bit Linux)
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Found 72 DICOM file(s)
INFO: stdout 2019-02-12T23:03:56.504069:Found 72 DICOM file(s)
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Warning: This diffusion series does not have a B0 (reference) volume
INFO: stdout 2019-02-12T23:03:56.504069:Warning: This diffusion series does not have a B0 (reference) volume
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Note: this appears to be a b=0+trace DWI; ADC/trace removal has been disabled.
INFO: stdout 2019-02-12T23:03:56.504069:Note: this appears to be a b=0+trace DWI; ADC/trace removal has been disabled.
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Warning: Volume 0 appears to be an ADC map (non-zero b-value with zero vector length)
INFO: stdout 2019-02-12T23:03:56.504069:Warning: Volume 0 appears to be an ADC map (non-zero b-value with zero vector length)
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Warning: Saving 1 DTI gradients. Validate vectors (image slice orientation not reported, e.g. 2001,100B).
INFO: stdout 2019-02-12T23:03:56.504069:Warning: Saving 1 DTI gradients. Validate vectors (image slice orientation not reported, e.g. 2001,100B).
190212-23:03:56,504 nipype.interface INFO:
stdout 2019-02-12T23:03:56.504069:Convert 72 DICOM as ./dwi (128x128x72x1)
INFO: stdout 2019-02-12T23:03:56.504069:Convert 72 DICOM as ./dwi (128x128x72x1)
190212-23:03:56,521 nipype.interface INFO:
stdout 2019-02-12T23:03:56.521709:compress: “/usr/bin/pigz” -n -f -6 “./dwi.nii”
INFO: stdout 2019-02-12T23:03:56.521709:compress: “/usr/bin/pigz” -n -f -6 “./dwi.nii”
190212-23:03:56,521 nipype.interface INFO:
stdout 2019-02-12T23:03:56.521709:Conversion required 0.070095 seconds (0.046532 for core code).
INFO: stdout 2019-02-12T23:03:56.521709:Conversion required 0.070095 seconds (0.046532 for core code).
190212-23:03:56,541 nipype.workflow INFO:
[Node] Finished “convert”.
INFO: [Node] Finished “convert”.
190212-23:03:56,553 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmetaphx1fn9m/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmetaphx1fn9m/embedder”.
190212-23:03:56,565 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
190212-23:03:56,570 nipype.workflow INFO:
[Node] Finished “embedder”.
INFO: [Node] Finished “embedder”.
INFO: Post-treating /base/Nifti/sub-001/ses-01/dwi/sub-001_ses-01_run-2_dwi.json file
INFO: Populating template files under /base/Nifti/
INFO: PROCESSING DONE: {‘subject’: ‘001’, ‘outdir’: ‘/base/Nifti/’, ‘session’: ‘01’}