Hi friends,
following issue: I’m following this tutorial to convert .dcm to BIDS: BIDS Tutorial Series: HeuDiConv Walkthrough | Stanford Center for Reproducible Neuroscience. I’m stuck on step 4, where I run HeuDiConv with heuristic.py:
docker run --rm -it -v C:/Users/pazlab/Documents/Max/MRIData/Pilot:/base nipy/heudiconv:latest -d /base/{subject}/*/*/*/*.dcm -o /base/Nifti/ -f /base/Nifti/code/heuristic.py -s "BE796" -c dcm2niix -b --overwrite
Running this, I get the following output:
C:\Users\pazlab>docker run --rm -it -v C:/Users/pazlab/Documents/Max/MRIData/Pilot:/base nipy/heudiconv:latest -d /base/{subject}////.dcm -o /base/Nifti/ -f /base/Nifti/code/heuristic.py -s “BE796” -c dcm2niix -b --overwrite
INFO: Running heudiconv version 0.9.0 latest 0.10.0
INFO: Need to process 1 study sessions
INFO: PROCESSING STARTS: {‘subject’: ‘BE796’, ‘outdir’: ‘/base/Nifti/’, ‘session’: None}
INFO: Processing 2104 dicoms
INFO: Analyzing 2104 dicoms
INFO: Generated sequence info for 14 studies with 2104 entries total
INFO: Doing conversion using dcm2niix
INFO: Lock 140494038703856 acquired on /base/Nifti/heudiconv.lock
INFO: Populating template files under /base/Nifti/
INFO: Lock 140494038703856 released on /base/Nifti/heudiconv.lock
INFO: PROCESSING DONE: {‘subject’: ‘BE796’, ‘outdir’: ‘/base/Nifti/’, ‘session’: None}
And in my Nifti folder, I get no .json outputs except my participants .json.
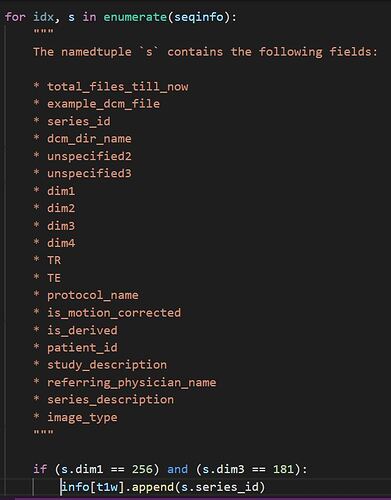
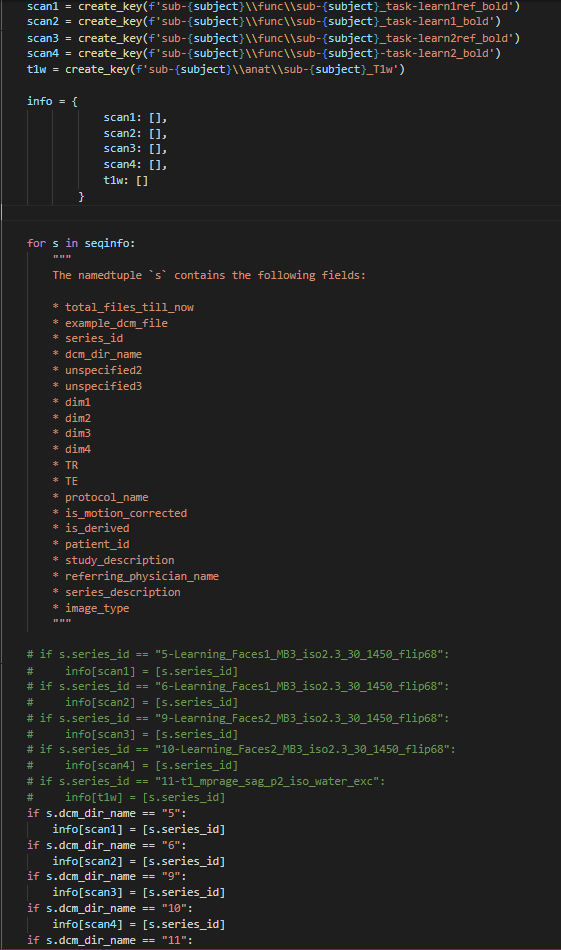
It looks like it’s not recognizing the identifiers I give in my dicominfo.tsv file but I’m not sure why, or at least for some other reason it seems like it doesn"t manage to read the files.
Here’s my code:
I’ve tried lots of different things, among them:
- Changed the file identifier (as you can see in the bracketed out part)
- Playing around with syntax (i.e. different backslashes etc., as you can see on top of the picture)
Not sure exactly what the issue is. Any leads welcome!
Thanks
Max
 Thanks for your reply! Tried playing around with the paths separators but it doesn’t seem to be the issue. I’ve been searching the heudiconv documentation on how to use the HEUDICONV_LOG_LEVEL=DEBUG setting, but don’t exactly understand how to use it - could you provide an example?
Thanks for your reply! Tried playing around with the paths separators but it doesn’t seem to be the issue. I’ve been searching the heudiconv documentation on how to use the HEUDICONV_LOG_LEVEL=DEBUG setting, but don’t exactly understand how to use it - could you provide an example?