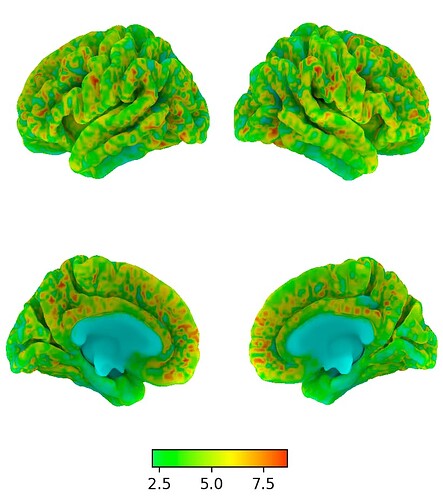
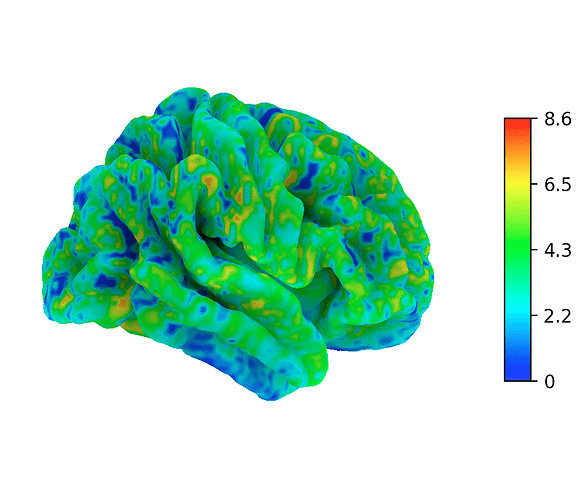
Here is a solution how to plot a non-symmetrical colormap or colorbar in nilearn, even though that is normally not supported by nilearn.
I have seen this question in a couple of topics: people asked how one can plot a colormap or colorbar with only positive (or only negative) values, without nilearn automatically re-arranging the zero- or centerpoint of the colorbar.
You can “trick” nilearn by using a combination of the same colormap, where you combine the colormap two times into one new colormap as follows. But see the script below.
import nilearn
import numpy as np
import cmasher as cmr
from nilearn import surface
from nilearn import plotting
from nilearn import datasets
# Initialization
color = "hsv_r"
range_cmap = cmr.get_sub_cmap(color, 0.3, 1)
# Custom colormap
from matplotlib import cm
from matplotlib.colors import ListedColormap, LinearSegmentedColormap
upper = cm.get_cmap(range_cmap, 999)
lower = cm.get_cmap(range_cmap, 999)
combine = np.vstack((upper(np.linspace(0, 1, 999)),
lower(np.linspace(0, 1, 999))))
custom_cmap = ListedColormap(combine, name="custom_map")
# Data
nifti = nilearn.image.load_img("load-your-nifti-file")
# Cortical mesh
fsaverage = datasets.fetch_surf_fsaverage(mesh="fsaverage")
# Sample the 3D data around each node of the mesh
texture = surface.vol_to_surf(nifti, fsaverage.pial_right)
# Plot
fig = plotting.plot_surf_stat_map(
surf_mesh=fsaverage.pial_right,
stat_map=texture,
bg_map=fsaverage.sulc_right,
bg_on_data=True,
alpha=1,
vmax=8.64,
threshold=False,
hemi='right',
title='Surface right hemisphere',
colorbar=True,
symmetric_cbar=False,
cmap=custom_cmap,
)
fig.show()
The result is that nilearn now uses the complete colormap that you specified above, instead of “re-centering” it. Of course, nilearn still trys to re-center the colormap, but since your chosen colormap is mirrored around the centerpoint, the code still catches the complete map, now displaying the full range even though all values are positive (or negative).
Even the plotted colorbar only shows the intended range, while in fact, the “real” colorbar consists of a combination that uses two times the same colormap.