Summary of what happened:
I try to use fmriprep-docker to run multi-echo data but the results did not combine the multi echos. I think that somewhere of my command need to be corrected but I have no idea right now.
Command used (and if a helper script was used, a link to the helper script or the command generated):
The config.json is as below:
{
"descriptions": [
{
"dataType": "anat",
"modalityLabel": "T1w",
"criteria": {
"SeriesDescription": "t1_mprage_sag_p2",
"ProtocolName": "t1_mprage_sag_p2",
"SeriesNumber": 2
}
},
{
"dataType": "func",
"modalityLabel": "bold",
"customLabels": "task-rest_run-01",
"criteria": {
"SeriesDescription": "REsting_3.0iso_conv_GRA2_MB3_ME4",
"ProtocolName": "REsting_3.0iso_conv_GRA2_MB3_ME4",
"ScanningSequence": "EP"
},
"sidecarChanges": {
"TaskName": "rest"
}
}
]
}
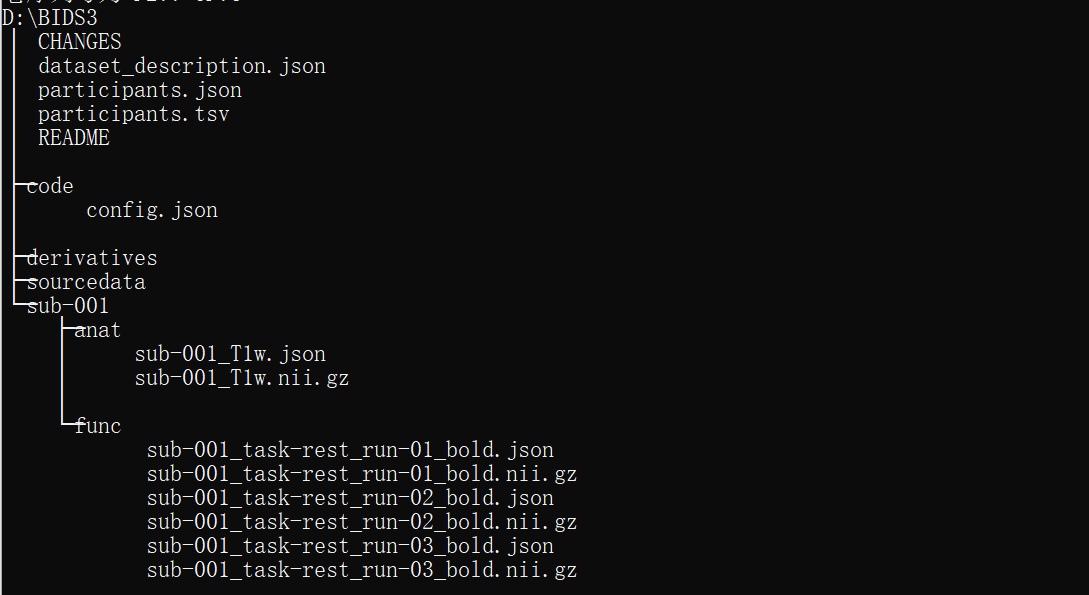
The command on PowerShell like:
fmriprep-docker D:\bids2 D:\bids2\derivatives participant --participant-label 001 --fs-license-file D:\FClearning\license.txt --fs-no-reconall
Version:
fmriprep:22.1.1
Environment (Docker, Singularity, custom installation):
Docker
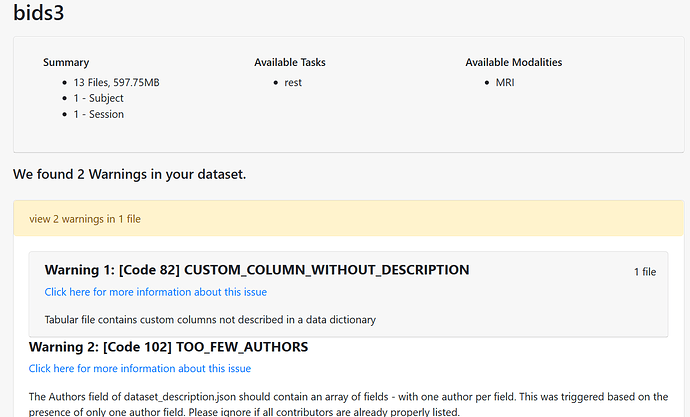
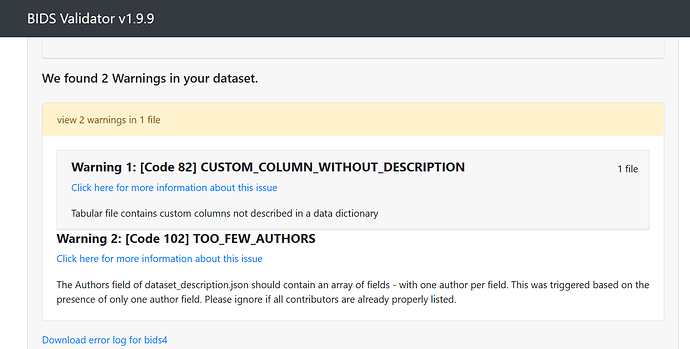
Data formatted according to a validatable standard? Please provide the output of the validator:
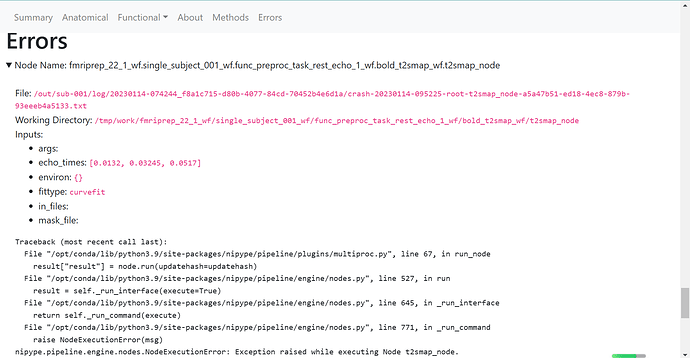
Relevant log outputs (up to 20 lines):
Here is the logs/CITATION.tex:
Results included in this manuscript come from preprocessing performed
using \emph{fMRIPrep} 22.1.1 (\citet{fmriprep1}; \citet{fmriprep2};
RRID:SCR\_016216), which is based on \emph{Nipype} 1.8.5
(\citet{nipype1}; \citet{nipype2}; RRID:SCR\_002502).
\begin{description}
\item[Anatomical data preprocessing]
A total of 1 T1-weighted (T1w) images were found within the input BIDS
dataset.The T1-weighted (T1w) image was corrected for intensity
non-uniformity (INU) with \texttt{N4BiasFieldCorrection} \citep{n4},
distributed with ANTs 2.3.3 \citep[RRID:SCR\_004757]{ants}, and used as
T1w-reference throughout the workflow. The T1w-reference was then
skull-stripped with a \emph{Nipype} implementation of the
\texttt{antsBrainExtraction.sh} workflow (from ANTs), using OASIS30ANTs
as target template. Brain tissue segmentation of cerebrospinal fluid
(CSF), white-matter (WM) and gray-matter (GM) was performed on the
brain-extracted T1w using \texttt{fast} \citep[FSL 6.0.5.1:57b01774,
RRID:SCR\_002823,][]{fsl_fast}. Volume-based spatial normalization to
one standard space (MNI152NLin2009cAsym) was performed through nonlinear
registration with \texttt{antsRegistration} (ANTs 2.3.3), using
brain-extracted versions of both T1w reference and the T1w template. The
following template was selected for spatial normalization: \emph{ICBM
152 Nonlinear Asymmetrical template version 2009c}
{[}\citet{mni152nlin2009casym}, RRID:SCR\_008796; TemplateFlow ID:
MNI152NLin2009cAsym{]}.
\item[Functional data preprocessing]
For each of the 3 BOLD runs found per subject (across all tasks and
sessions), the following preprocessing was performed. First, a reference
volume and its skull-stripped version were generated using a custom
methodology of \emph{fMRIPrep}. Head-motion parameters with respect to
the BOLD reference (transformation matrices, and six corresponding
rotation and translation parameters) are estimated before any
spatiotemporal filtering using \texttt{mcflirt} \citep[FSL
6.0.5.1:57b01774,][]{mcflirt}. BOLD runs were slice-time corrected to
0.694s (0.5 of slice acquisition range 0s-1.39s) using \texttt{3dTshift}
from AFNI \citep[RRID:SCR\_005927]{afni}. The BOLD time-series
(including slice-timing correction when applied) were resampled onto
their original, native space by applying the transforms to correct for
head-motion. These resampled BOLD time-series will be referred to as
\emph{preprocessed BOLD in original space}, or just \emph{preprocessed
BOLD}. The BOLD reference was then co-registered to the T1w reference
using \texttt{mri\_coreg} (FreeSurfer) followed by \texttt{flirt}
\citep[FSL 6.0.5.1:57b01774,][]{flirt} with the boundary-based
registration \citep{bbr} cost-function. Co-registration was configured
with six degrees of freedom. Several confounding time-series were
calculated based on the \emph{preprocessed BOLD}: framewise displacement
(FD), DVARS and three region-wise global signals. FD was computed using
two formulations following Power (absolute sum of relative motions,
\citet{power_fd_dvars}) and Jenkinson (relative root mean square
displacement between affines, \citet{mcflirt}). FD and DVARS are
calculated for each functional run, both using their implementations in
\emph{Nipype} \citep[following the definitions by][]{power_fd_dvars}.
The three global signals are extracted within the CSF, the WM, and the
whole-brain masks. Additionally, a set of physiological regressors were
extracted to allow for component-based noise correction
\citep[\emph{CompCor},][]{compcor}. Principal components are estimated
after high-pass filtering the \emph{preprocessed BOLD} time-series
(using a discrete cosine filter with 128s cut-off) for the two
\emph{CompCor} variants: temporal (tCompCor) and anatomical (aCompCor).
tCompCor components are then calculated from the top 2\% variable voxels
within the brain mask. For aCompCor, three probabilistic masks (CSF, WM
and combined CSF+WM) are generated in anatomical space. The
implementation differs from that of Behzadi et al.~in that instead of
eroding the masks by 2 pixels on BOLD space, a mask of pixels that
likely contain a volume fraction of GM is subtracted from the aCompCor
masks. This mask is obtained by thresholding the corresponding partial
volume map at 0.05, and it ensures components are not extracted from
voxels containing a minimal fraction of GM. Finally, these masks are
resampled into BOLD space and binarized by thresholding at 0.99 (as in
the original implementation). Components are also calculated separately
within the WM and CSF masks. For each CompCor decomposition, the
\emph{k} components with the largest singular values are retained, such
that the retained components' time series are sufficient to explain 50
percent of variance across the nuisance mask (CSF, WM, combined, or
temporal). The remaining components are dropped from consideration. The
head-motion estimates calculated in the correction step were also placed
within the corresponding confounds file. The confound time series
derived from head motion estimates and global signals were expanded with
the inclusion of temporal derivatives and quadratic terms for each
\citep{confounds_satterthwaite_2013}. Frames that exceeded a threshold
of 0.5 mm FD or 1.5 standardized DVARS were annotated as motion
outliers. Additional nuisance timeseries are calculated by means of
principal components analysis of the signal found within a thin band
(\emph{crown}) of voxels around the edge of the brain, as proposed by
\citep{patriat_improved_2017}. The BOLD time-series were resampled into
standard space, generating a \emph{preprocessed BOLD run in
MNI152NLin2009cAsym space}. First, a reference volume and its
skull-stripped version were generated using a custom methodology of
\emph{fMRIPrep}. All resamplings can be performed with \emph{a single
interpolation step} by composing all the pertinent transformations
(i.e.~head-motion transform matrices, susceptibility distortion
correction when available, and co-registrations to anatomical and output
spaces). Gridded (volumetric) resamplings were performed using
\texttt{antsApplyTransforms} (ANTs), configured with Lanczos
interpolation to minimize the smoothing effects of other kernels
\citep{lanczos}. Non-gridded (surface) resamplings were performed using
\texttt{mri\_vol2surf} (FreeSurfer).
\end{description}
Screenshots / relevant information:
Hello all,
I am new to use fMRIPrep, and I have a confusing question about how to process multi echo data on fMRIPrep. What I have learnt is that first using dcm2bids to transform the raw multi echo data to BIDS, and then using fmriprep-docker to run the pipline(my PC is based on Windows).I don’t know if I understand right?
What’s more, I am not sure how to write the acquired config.json for multi echo data, since there seems no tutorials about it. I just try to use the basic command to run multi-echo data(since I read somewhere that fMRIPrep will automatically combine the echos) but the results seems to process the echo data seperately (just like single scho).
My command on PowerShell is like below,and I don’t know if it is right:
fmriprep-docker D:\bids2 D:\bids2\derivatives participant --participant-label 001 --fs-license-file D:\FClearning\license.txt --fs-no-reconall
I would really appreciate it if I can get any reply! Thanks a lot!
Andrea