I am trying to overlay a normalized anatomical image to the normalized functional image and I need to see the anatomical image as an edge overlay on the functional image.
How can it be done using FSLeyes or AFNI or any other neuroimaging viewer?
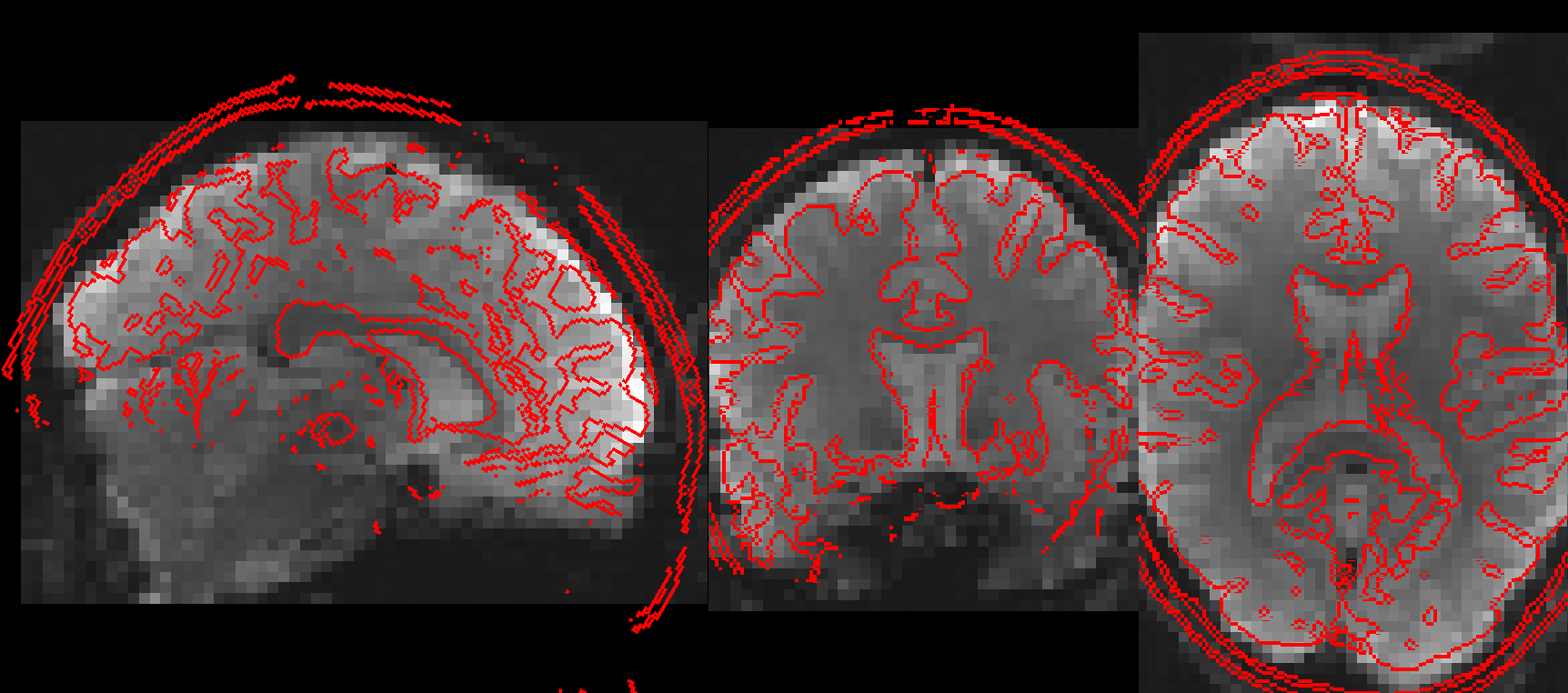
AFNI has two main edge-estimation programs: 3dedge3 and 3dedgedog.
You could create a dataset of edges, and load that in AFNI to view and/or save images.
I’ll also note that showing edges of an anatomical over an EPI is definitely useful for showing quality of alignment; that is what AFNI’s afni_proc.py uses for the EPI-anatomical (and anatomical-template) alignment checks for its QC HTML, as demonstrated here.
Specifically, the script/program that takes the EPI as underlay and anatomical as overlay, and calculates the edges internally and then saves a montage image for each of the major slice planes, is demonstrated here:
https://afni.nimh.nih.gov/pub/dist/doc/htmldoc/tutorials/auto_image/auto_%40chauffeur_afni.html#ex-8-check-alignment-with-edge-view
That might be particularly useful. Having that program to do the snapshotting means you can systematically view the outputs, which is convenient.
–pt
Hi,
you can easily do that with nilearn; See
Best,
Bertrand
Hi Bertrand
Thanks for your response. I tried Nilearn and it accepts 3D images but I have 4D functional images.
Thanks ptaylor!
The link was very helpful and did the job.
Indeed you may want to pick an image (or loop over images) with nilearn.image.index_img function or take some statistical summary such as the mean, for display.
HTH,
Bertrand
For what it’s worth, you can do this in FSLeyes by doing the following:
- Selecting the top image (the one you want to see as edges) and opening the overlay display panel (the gear button at top-left).
- changing the Overlay data type to to 3D/4D mask image
- Turning on Show outline only
- Adjusting the Threshold until you are happy (25th and 75th percentiles of the image data range often work well)
- Setting the outline width/colour/interpolation as desired
You can do this from the command line with a command such as:
fsleyes func.nii.gz struct.nii.gz -ot mask -mc 1 0 0 -o -t 100 600