I’m running nilearn’s decoder…
generic_mask_filepath = '.../spm12/canonical/MNI152_T1_1mm_brain_mask.nii'
decoder = Decoder(estimator = 'svc', mask = generic_mask_filepath, standardize= True)
prediction = decoder.fit(subj_data, predict_var)
and receiving a resampling warning:
.conda/envs/neuralsignature/lib/python3.8/site-packages/nilearn/image/resampling.py:594: RuntimeWarning: NaNs or infinite values are present in the data passed to resample. This is a bad thing as they make resampling ill-defined and much slower.
_resample_one_img(data[all_img + ind], A, b, target_shape,
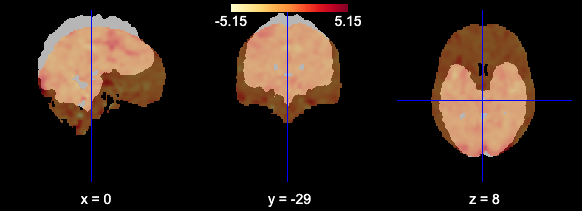
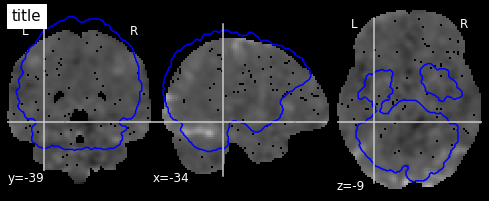
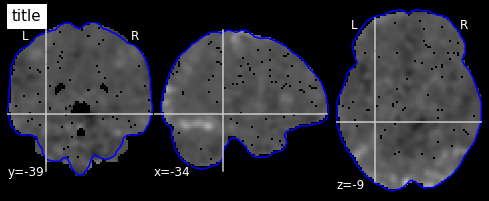
I worry this is because the mask I’m using and the fMRI data are not being aligned correctly. When I plot using the plotting function they appear to be misaligned:
mask_generic = image.load_img(generic_mask_filepath)
plotting.view_img(image.mean_img(subj_data),mask=mask_generic,opacity=0.7,cmap='YlOrRd')
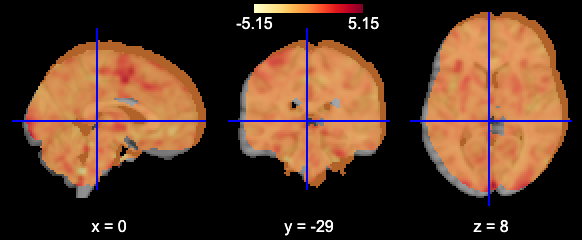
And alignment is something we need to be concerned about because the images don’t have the same affine:
print(image.mean_img(subj_data).affine)
[[ 2. 0. 0. -96.]
[ 0. 2. 0. -132.]
[ 0. 0. 2. -78.]
[ 0. 0. 0. 1.]]
print(mask_generic.affine)
[[ -1. 0. 0. 90.]
[ 0. 1. 0. -126.]
[ 0. 0. 1. -72.]
[ 0. 0. 0. 1.]]
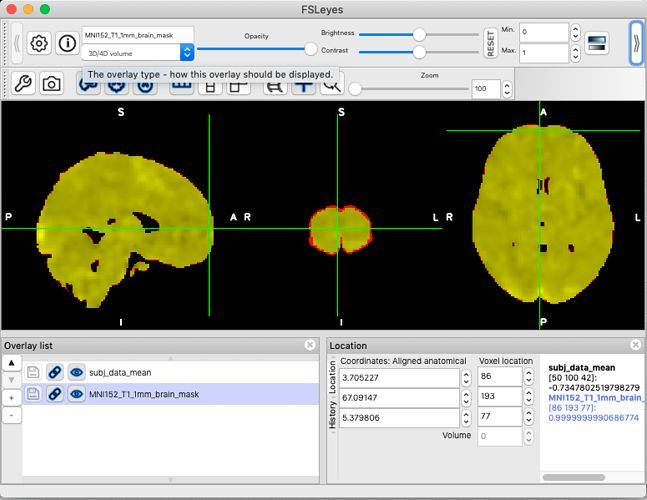
However, when loading in FSLeyes, the two images appear to be correctly aligned (albeit the subject data looks a little more tightly trimmed than the standard image).
(as a new poster I can’t insert more than one media item in a post, so the FSLeyes screenshot is in a reply below instead)
Several questions:
(1) Is there an alignment problem during the decoding (I’m not worried about alignment in visualization; it just seems possibly diagnostic), as there is during the image viewing?
(2) If so, it the cause of the warning message I’m seeing?
(3) …and how could I address the alignment problem?
(4) If there’s no alignment problem during decoding, how can I be confident about that?