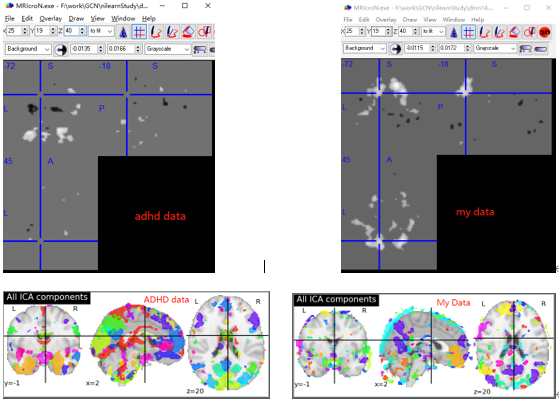
Hello, I also need to use ICA as a single object extraction network. But in the process of using it, I found some differences. An ADHD data and my data are shown below(Both use an object,one adhd data and one rsfMRI data). My data shows that there is something wrong with the results. Is the result normal? Is it my problem of data preprocessing? Or is there a problem with some codes? Is the extracted result correct?
codes:
num = 40
adhd_data = datasets.fetch_adhd(n_subjects=num)
func = adhd_data.func[0]
# func = r'I:\TenDrugDataNoBrain\Drug01_brain.nii' # my data
canica = decomposition.CanICA(n_components=20, mask_strategy='background')
#
# # # func_filenames = rest_dataset.func
canica.fit(func)
#Retrieving the components
components = canica.components_
# canica_components_img = canica.components_img_
#Using a masker to project into the 3D space
components_img = canica.masker_.inverse_transform(components)
canica_components_img = image.index_img(components_img, 9)
#Plotting the default mode network (DMN) without region extraction
plotting.plot_stat_map(image.index_img(components_img, 9), title='DMN')
plotting.show()
# Plotting all the components
plotting.plot_prob_atlas(components_img, title='All ICA components')
plotting.show()
canica_components_img.to_filename('dmn13.nii.gz')