Hi,
I have a difficult problem. The data I have was acquired from the right side of the brain obliquely and only contains part of the FOV, so my raw fMRI images have a deflection angle. After I used the AFNI 3dresample, the image has improved in resolution but my image has no more deflection and seems to have become horizontal, causing me to be unable to align with the original T1. Do you know how to solve this problem please? Thank you very much for your help! ![]()
Hi @Shuyue_Fu ,
Welcome to Neurostars!
How did you use with 3dresample? Could you paste the command line you use and what output was the command giving you?
Thank you for your reply! Yes. I used AFNI’s 3dresample. The code is as follows:
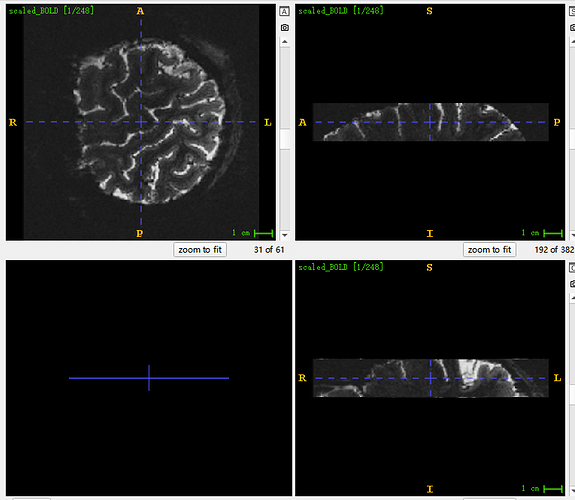
This is the image I visualized with ITK-snap. You can see in the lower left corner that the original image has a deflection Angle. But when I upsampled it, it became horizontal, so it couldn’t register with the original T1 image. Here is my upsampled image, which has tripled the voxel resolution.
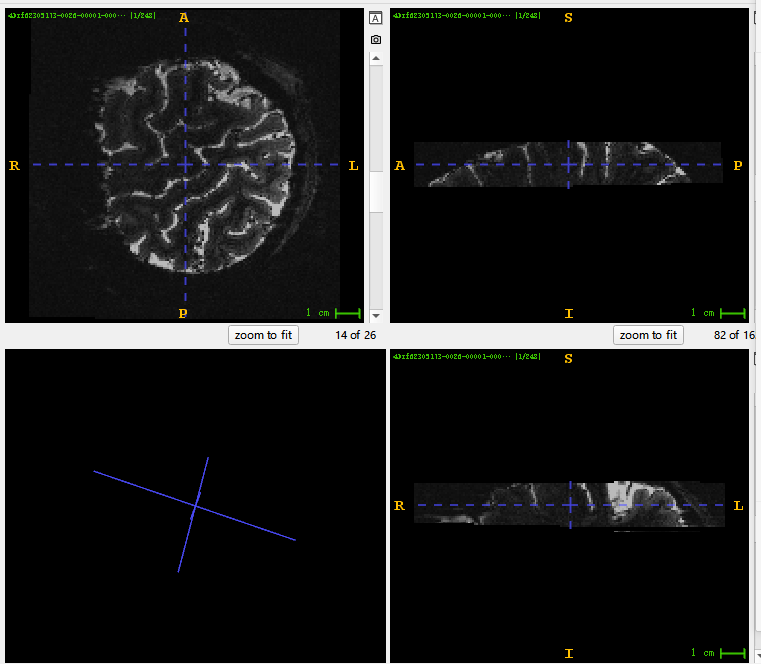
Sorry, new users can only paste one image, so I’ll add the image here. This is the original picture. You can see the deflection Angle in the lower left corner
3dresample removes that obliquity information from the header to make it cardinal. Instead of using 3dresample, you can use 3dWarp to simultaneously move the dataset to the anatomical dataset grid and resample to a higher resolution voxel size.
3dWarp -card2oblique myanat.nii.gz -newgrid 1.5 -prefix cardEPI.nii.gz myepi.nii.gz
Moving among different software might be a little confusing depending on the data. The AFNI GUI doesn’t use the obliquity information for display, only the closest cardinal orientation, and the results from that 3dWarp command will appear correct within AFNI. If the original anatomical dataset is also oblique, the ITKsnap viewer will not match. One solution is to “deoblique” a copy of everything with
cp dset.nii.gz copy.nii.gz
3drefit -deoblique copy.nii.gz
That changes the header of the dataset without resampling. Alternatively, you can deoblique with 3dWarp to put both the EPI and anatomical datasets into a cardinal grid.
3dWarp -deoblique -prefix anat_deob.nii.gz anat.nii.gz
3dWarp -deoblique -prefix epi_deob.nii.gz -newgrid 1.5 epi.nii.gz
For EPI datasets, be sure to do slice timing correction before doing any spatial transformations like resampling or warping. Programs like afni_proc.py and align_epi_anat.py will handle all these operations. Partial coverage data like this may require one of the -partial_… options in align_epi_anat.py. The -master_… options can control the grid of the output for align_epi_anat.py.
For more information,
Thank you for your answer. Your answer was very detailed and solved my question. Sincerely express your gratitude again! ![]()