Hello,
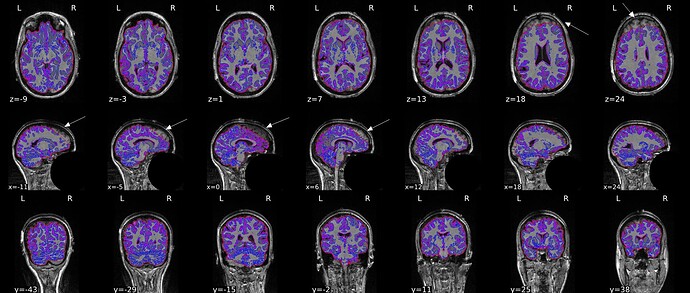
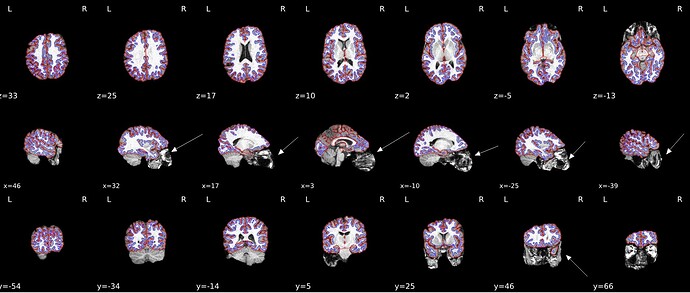
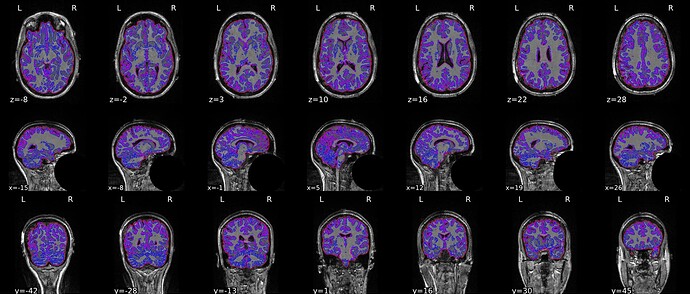
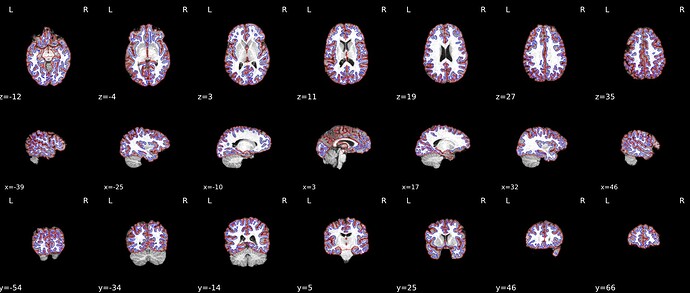
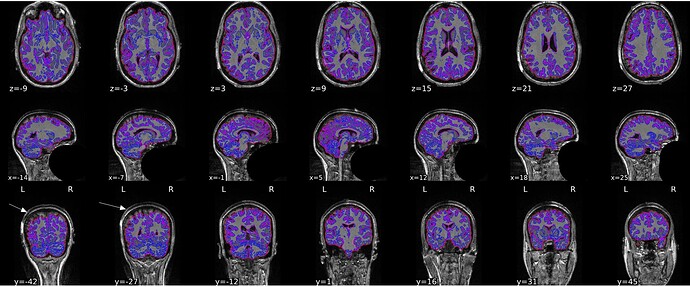
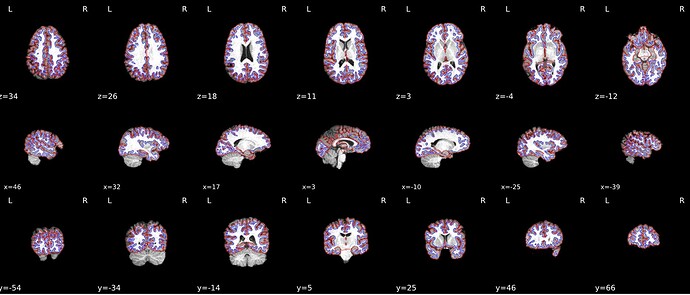
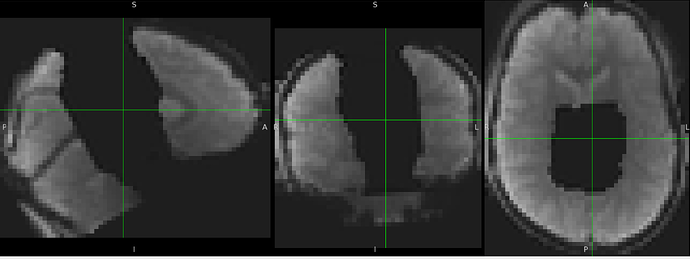
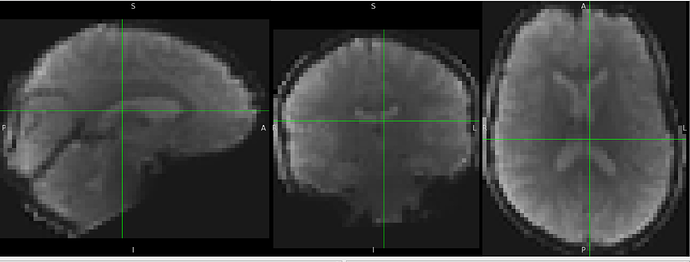
I got inaccurate brain mask estimation when enabling --use-syn-sdc and there are some wired ghost artifacts in the brain image. When--use-syn-sdc is not used, everything looks fine. Do you know what’s wrong here? Please find below the snapshots of HTML reports and fmriprep codes. Thanks.
* fMRIPrep version: 21.0.2
* fMRIPrep command: `/opt/conda/bin/fmriprep -w /DATA/fmriprep_work --participant-label 1111 --bids-filter-file /OUTPUT/bids_filters.json --output-spaces MNI152NLin2009cAsym MNI152NLin6Asym T1w fsnative fsaverage --output-layout bids --notrack --skip_bids_validation --use-syn-sdc --write-graph --omp-nthreads 8 --nprocs 16 --mem_mb 65536 --resource-monitor /DATA/EEGfMRI_prep /DATA/EEGfMRI_prep/derivatives/fmriprep participant`
* Date preprocessed: 2022-06-25 20:19:06 +0000
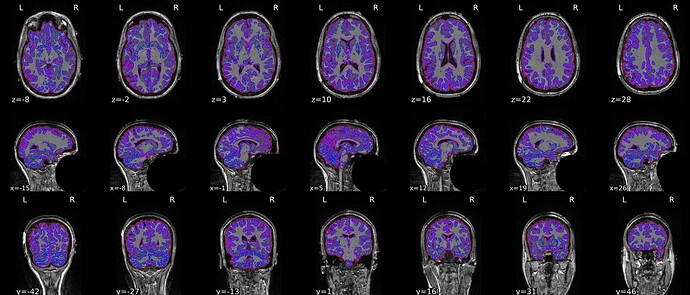
Brain mask and brain tissue segmentation of the T1w
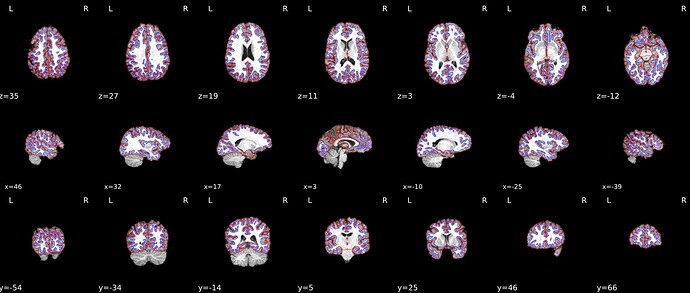
Surface reconstruction
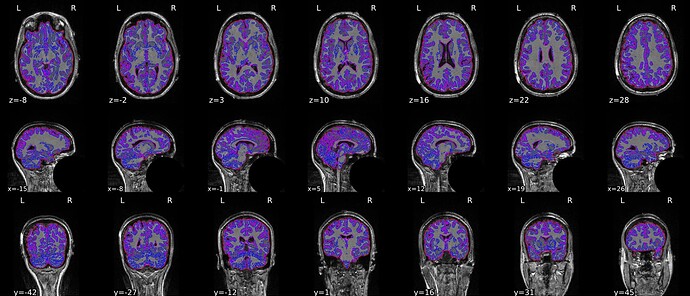
When --use-syn-sdc is not used, both brain mask and brain image look fine.
* fMRIPrep version: 21.0.2
* fMRIPrep command: `/opt/conda/bin/fmriprep -w /DATA/fmriprep_work --participant-label 1111 --bids-filter-file /OUTPUT/bids_filters.json --output-spaces MNI152NLin2009cAsym MNI152NLin6Asym T1w fsnative fsaverage --output-layout bids --notrack --skip_bids_validation --write-graph --omp-nthreads 8 --nprocs 16 --mem_mb 65536 --resource-monitor /DATA/EEGfMRI_prep /DATA/EEGfMRI_prep/derivatives/fmriprep participant`
* Date preprocessed: 2022-06-26 15:14:37 +0000
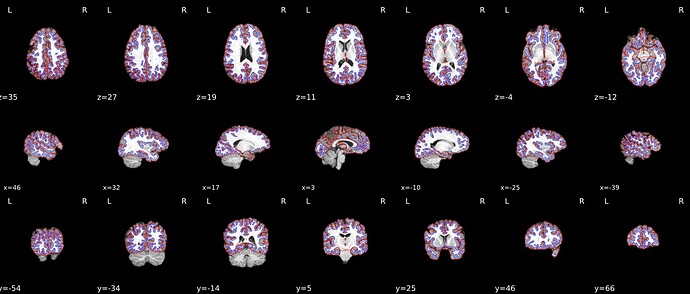
A previous fmriprep version also worked fine without --use-syn-sdc
* fMRIPrep version: 20.2.5
* fMRIPrep command: `/usr/local/miniconda/bin/fmriprep -w /DATA/fmriprep_work --participant-label 1111 --bids-filter-file /OUTPUT/bids_filters.json --output-spaces MNI152NLin2009cAsym MNI152NLin6Asym T1w fsnative fsaverage --output-layout bids --notrack --skip_bids_validation --write-graph --omp-nthreads 8 --nprocs 16 --mem_mb 65536 --resource-monitor /DATA/EEGfMRI_prep /DATA/EEGfMRI_prep/derivatives/fmriprep participant`
* Date preprocessed: 2022-06-24 21:51:26 +0000