Summary of what happened:
Dear all,
We encountered unexpected output from fMRIPrep when processing task-based fMRI data acquired on a Siemens 3 T Cima.X scanner. The dataset includes pairs of spin-echo fieldmaps with reversed phase-encoding directions for distortion correction.
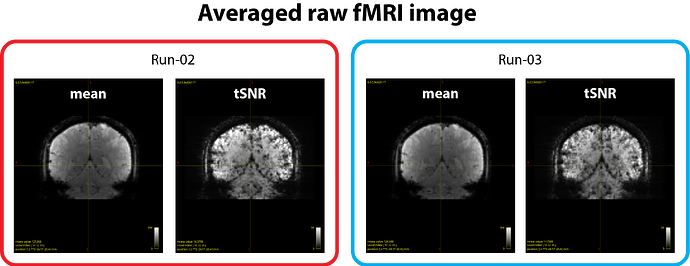
During acquisition, we did not use prescan normalization, so the raw fMRI data naturally show an intensity bias, as expected (see figure 1 below).
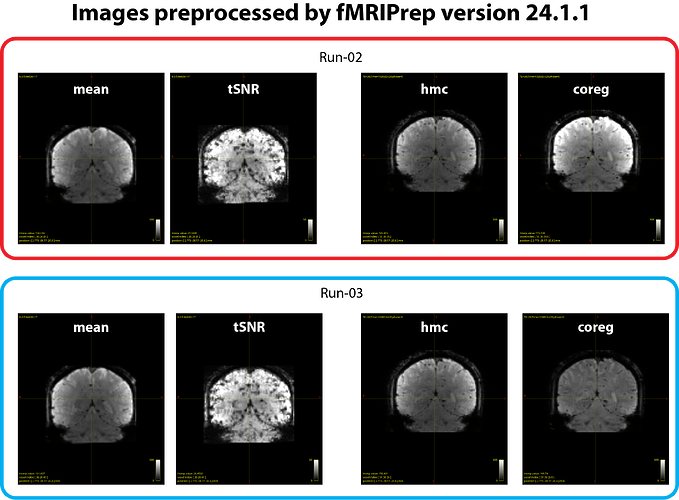
The issue is that the image intensity of the *_desc-coreg_boldref.nii.gz files is inconsistent across runs. As we understand, this image is used for coregistration to the T1-weighted image, and a bias-field correction is typically applied to improve alignment. However, this correction seems to succeed in some runs (e.g. run-03 in our case) but not others (e.g. run-02 in our case), resulting in marked intensity differences across runs. We have attached example images illustrating the issue (see figure 2 below). This phenomenon occurred with fMRIPrep v24.1.1, and runs showing this inconsistency also yield weaker or absent BOLD z-score signals in the GLM results.
We verified that the raw and other preprocessed images appear consistent across runs:
- Averaged raw fMRI images show comparable intensity bias levels (see figure 1).
- tSNR maps look similar across runs (see figure 1 and 2).
*_desc-hmc_boldref.nii.gzimages (generated just before the coregistration step) are also consistent (see figure 2).
Thus, the inconsistency seems to arise at the *_desc-coreg_boldref.nii.gz step.
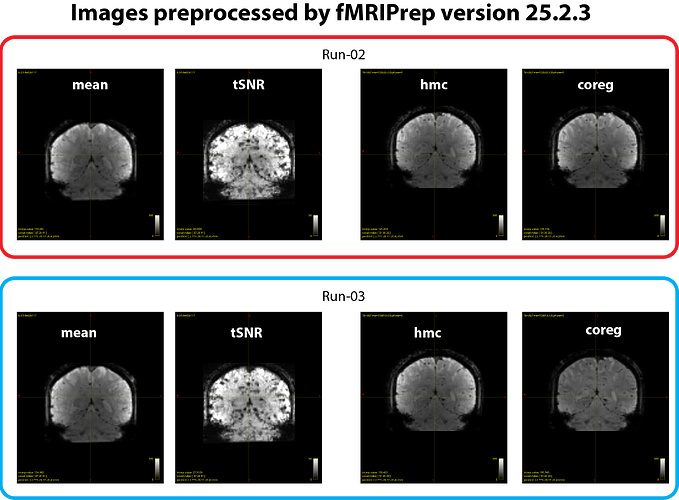
We suspected whether updating the fMRIPrep version would help. When we rerun the same analysis using ver 25.2.3, we found that a degree of inter-run inconsistency in *_desc-coreg_boldref.nii.gz images became much less compared with output from ver 24.1.1. However, perhaps some of the differences may still exist in this case, although it looks much better (see figure 3 below).
We are unsure where this issue originates. Perhaps, there is some instability in the step for applying intensity bias correction for *_desc-coreg_boldref.nii.gz images. Alternatively, there may be issues in applying the susceptibility-induced correction to this image.
We would like to confirm whether this behavior is expected or if it could indicate a preprocessing bug in earlier versions of fMRIPrep. Could you advise on whether any specific settings or tests are recommended to ensure consistent bias correction across runs?
Thank you very much for your time and assistance.
Command used (and if a helper script was used, a link to the helper script or the command generated):
Code section for preprocessing
#!/bin/bash
.
.
.
apptainer run \
--cleanenv \
-B "${BIDS_DIR}:/bids:ro" \
-B "${OUTPUT_DIR}:/out" \
-B "${FSLicense}:/fslicense.txt:ro" \
-B "${WORK_LOCAL}:/work" \
-B "${BIDSDB_LOCAL}:/bidsdb" \
-B "${FREESURFER_DIR_LOCAL}:/fsdir" \
-B "${TMPDIR}:${TMPDIR}" \
"${APPTAINER_IMAGE}" \
/bids /out participant \
--participant-label "${sub_ID}" \
--fs-license-file /fslicense.txt \
--fs-subjects-dir /fsdir \
--work-dir /work \
--bids-database-dir /bidsdb \
--nthreads "${NTHREADS}" \
--omp-nthreads "${OMP_NTHREADS}" \
--mem_mb "${MEMMB}" \
--output-spaces T1w:res-native \
--skip-bids-validation \
--resource-monitor
Version:
fMRIPrep v24.1.1 and fMRIPrep v25.2.3
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
The file structure was confirmed to be BIDS-valid using the online BIDS Validator.
The IntendedFor field was manually added to the AP and PA fieldmap JSON files using the following format:
ses-*/func/sub-*_ses-*_task-fMRI_run-*_bold.nii.gz
Susceptibility distortion correction was successfully applied to all functional images, as confirmed by visual inspection of the fMRIPrep reports.
Screenshots / relevant information:
Operaing system: Ubuntu 24.04.2 LTS