Hi all,
I am running into 1) a problem when using Nilearn’s SecondLevelModel to statistically test different contrasts specified by my design matrix. When I plot the statistical maps I, most of their significant z-scores are located outside the brain/mask picture of the Nilearn plotting functions. And 2) I am confused on what the mask_img argument in SecondLevelModel does exactly, and whether I should be using this. I will explain both topics in a bit more depth below.
**1. Plotting contrasts **
As input to my second level model, I have for each participant the z-transformed correlation coefficients of the amygdala with all other voxels in the brain (executed in a similar fashion as Producing single subject maps of seed-to-voxel correlation - Nilearn).
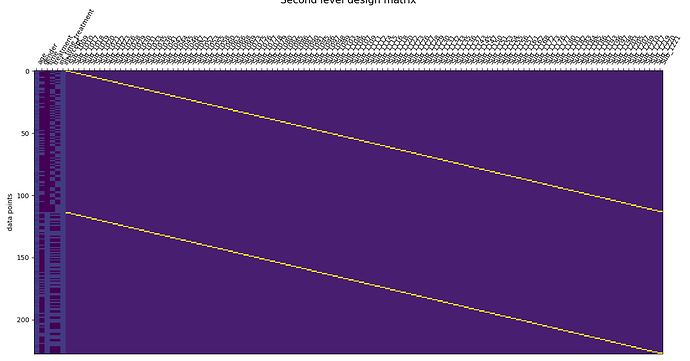
The design matrix in my second level model consists of the following columns: age & gender (covariates), time (baseline and follow-up), treatment (2 levels), intercept, and individual intercepts to account for repeated measures. I include a picture of my design matrix below.
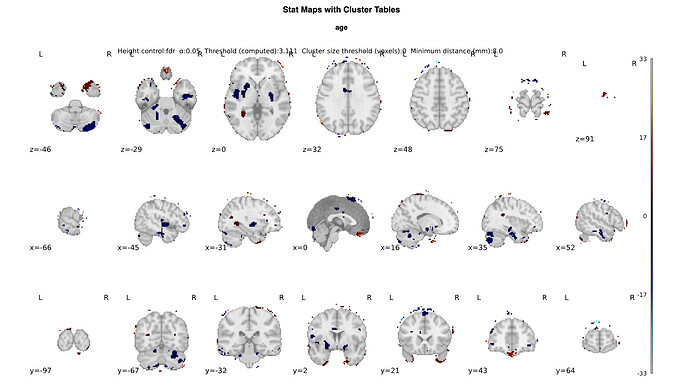
When I am interested in the effect of age on functional connectivity let’s say, I will run the following: left_stat_map_age = left_model.compute_contrast(‘age’, output_type=‘z_score’)
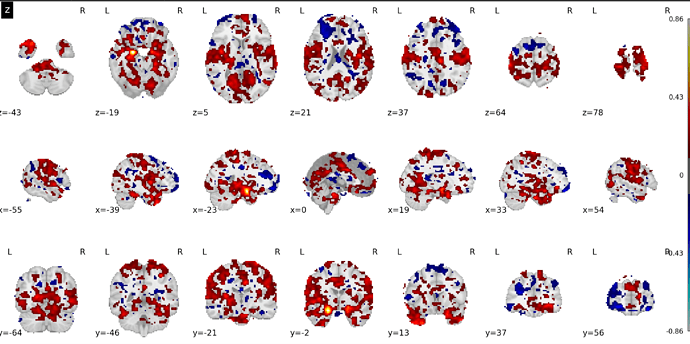
This will show the effect of age, keeping the effect of all other variables in my model constant. Now, the problem arises when I plot this contrast with a plotting function of Nilearn:
As you can see, the location of many ‘voxels’ that survive the fdr-correction have a problematic location: they are outside the brain image/mask within the plotting function. This data is not random however, and you can see that it distributed in a circular shape around the brain, as if that should be the outer edge of the brain image/mask. I have a difficult time finding out where this problem comes from. So any help is very appreciated. Do you have any ideas what could be the problem in this sort of problem?
2. argument mask_img
One solution I tried ties into my second question: What is the use of the argument mask_img in the SecondLevelModel exactly, and should I use this argument?
In the code for my first level correlation maps, I have already used the specific brain mask that fMRIPREP gives as output (in the same MNI space as my functional data - MNI152NLin2009cAsym). I have check this brain masks, and it fits very well on my preprocessed functional data from which the correlation maps are produced.
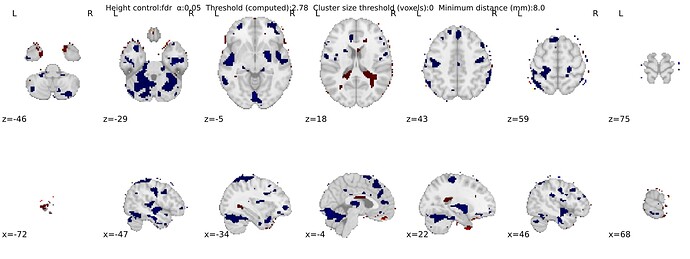
In this sense, the functional data as input into the SecondLevelModel is already masked. I did try to specify mask_img in this function as one fMRIPREP brain_mask nii.gz file (from a random participant), and this had an effect on the statistical maps: less severe finding of significance outside the brain - but still apparent:
To conclude, does anyone have an idea what could be going wrong - in what step of processing an error can occur to give rise to this distorted second level statistical output when I plot it. And second, how and when do you use the mask_img argument in the second level model. Is it a good idea to use this at the second level, or is that problematic?
Thanks a lot.