Hello Everyone,
For my research work I have been trying to learn DTI analysis. Since this is completely new, I am struggling to learn the software and solve any problem while I am stuck. On the top of that it is a animal brain. So using FSL causing problem (tbss analysis) and. now I am trying to learn DSI Studio and FSL more. Could you suggest some tutorial or lectures that would be helpful for me. So far, I have a very little knowledge about these.
Another question is that since it is a animal brain, can I skip registration steps and all that. I have extracted brain T1 images by hand using AFNI and thus the outline is not that good. Again, I have RD, AD map which is very unclear. Problem is that the T1 and RD maps have different resolution. if i create an ROI mask from T1 and resample it on RD map,is that correct. How do I know the value of RD or avg RD for that whole mask or region, since there are going to thousand of voxel there.
Thank you!
Hi @Sanjida,
Which animal? Some animals have more resources than others.
Perhaps look here: FSL Diffusion Toolbox Practical
Probably not - but what you registration steps you do/do not need to do is based on your analysis goals, not the species.
This article might help explain the concepts better: An introduction to diffusion tensor image analysis - PMC
I also like this page: How to interpret dMRI metrics | DSI Studio Documentation
This depends on what your analysis goals are. What you are defining as an ROI is important. Averaging within ROIs is a common approach though, but you’d have to be certain that your ROI is only white or gray matter, not some combination of brain compartments.
Best,
Steven
Howdy-
Re skullstripping the T1w images: I’m not sure what that would be done by hand. Why not use AFNI’s @animal_warper, for example? We have applied it to macaques, marmosets, canines and rodents, and it works across all of those species (and surely others). There are some demo datasets within AFNI for scripting with both @animal_warper and afni_proc.py to process macaque data:
- run
@Install_MACAQUE_DEMOfor the task demo - run
@Install_MACAQUE_DEMO_RESTfor the rest demo
… but again, those should apply across various species pretty directly.
–pt
Hi,
Thank you for your response. The one I am trying to do is piglets brain. I will try to see that how to do that the one you suggested.
OK, that sounds good.
Please let me know how it goes. I’m also curious, what piglet MRI template are you using? Does it have good definition/contrast?
–pt
Hi Steven,
Thank you so much. I have read and tried to do the TBSS analysis before but for the tbss_2_reg it did not work. The animal brain a piglet’s brain. And as a template I tried or FA template I tried to use one that I found online and it’s from UIUC.
Do you know if someone has run the same tbss commands for any rat on animal brain and how they have changed to make it work?
The FA map I created from MRTrix and the atlas I got from UIUC look bit different with their orientation like the sagittal axial are swapped and the rotation is different as well.
Do you that might be one of the reason or FSL registration should take care of that thing.
I am sorry to ask a very basic question in case I do not have any wrong idea. What does the registration for anything in FSL does actually? It put all the images in the same parameter, right? Or when we say we want to register our T1 and FA what it is supposed to do.
Thank you so much for being so helpful. I really appreciate. I am sorry I am lacking knowledge now since I just started learning but I will do my best.
Best,
Sanjida
Hi @Sanjida,
These article might provide clues: Miniature pig model of human adolescent brain white matter development - PMC , https://www.sciencedirect.com/science/article/pii/S016502702100042X, Maternal Dietary Choline Status Influences Brain Gray and White Matter Development in Young Pigs - PubMed
It depends on the analysis. For TBSS, for example, everything needs to be in the same space and aligned to each other, since tests are run on each segment of a single white matter skeleton. It is analogous to voxel-wise whole brain in fMRI research, but restricted only to white matter.
This is done to align DWI and T1 images. This can be useful for getting DWI images into additional spaces, as one can multiply the DWI–>T1 transformation with T1–>Otherspace transformation to get DWI–>Otherspace. Of course, this can also be inverted to bring Otherspace–>DWI.
Best,
Steven
Hi,
I tried seeing the commands and sorry I got little bit confused. does @animal_warper does some nonlinear wrap and skull stripping together and we need to have an atlas for that. I went through the scripts part for macaques to understand and did not really understand about which part i need to follow for the skull stripping. For example I have the nifti file of the T1 images with skull. If I just want to skull Stripp that file how shoul i implement that @animl_warper command. Can I do that.
Thank you so much.
Howdy-
Using @animal_warper requires having a reference template, but associated atlases and/or segmentations (i.e., “follower datasets” in the standard space) are optional. Indeed, it does perform both skullstripping and nonlinear alignment to the template, as those two processes feed off of each other.
Here is an example of running it with just an input anatomical (replace DSET_ANAT with your actual file name) and a reference template; I will assume that your reference template (replace DSET_TEMPLATE with that name, below) has no skull on it, so it can be used as its own mask:
@animal_warper \
-input DSET_ANAT \
-input_abbrev subj_anat \
-base DSET_TEMPLATE \
-base_abbrev tmplt \
-outdir odir_aw \
-ok_to_exist
If your DSET_TEMPLATE is not masked, then add in an option -skullstrip DSET_TEMPLATE_MASK with a mask for the standard space brain. I left the “abbrev” options in there, because it is often useful to have abbreviations for output file names—file names nowadays can be veeery long. The “-ok_to_exist” is handy, in case you re-run the processing later adding in followers, say.
How does that seem?
–pt
Hello,
Thank you so much. Sadly, I don’t have the pig template. Can you tell me where I can find a piglets’ template. Right now, I just have the raw images of T1(with skull) and without skull T1 (DONE BY HAND) and the FA map I got from MRtrix. I could find an atlas from UIUC (Pig Imaging Group website) but while I was trying to do the TBSS analysis it did not work. I am assuming, is it because of the different orientation of the pig atlas and our piglets’ images. just to double check if I understood correctly, I can run this commands in the terminal of linux
@animal_warper
-input DSET_ANAT (T1 image with skull the one i am trying to process)
-input_abbrev subj_anat ( output of that DSET_ANAT)
-base DSET_TEMPLATE ( pig template after i found one)
-base_abbrev tmplt (output of that pig tempate)
-outdir odir_aw ( output will be saved in this directory)
-ok_to_exist
I am sorry for asking such basic question. Since I am a new learner to all this. I am learing how to work in linux as wee. I hope you will understand. Thank you so much for your help and patience.
Best,
Sanjida
Howdy-
Re. finding a pig template, I don’t have one that I have used before. I searched a bit, and there appears to be one publicly available one from Chang et al. (2020) here:
https://www.nitrc.org/projects/micropig_brain
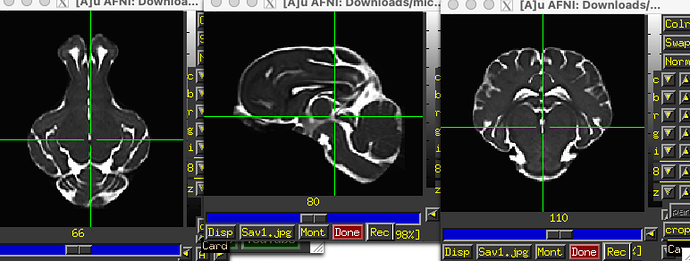
The T2w volume there looks the cleanest (the T1 is pretty noisy). I’m not sure why there is so much empty space vertically, but then probably over-tightness along the AP axis. Also, annoyingly, there is obliquity in the header. It really should not have been constructed in that way. To clean up a few things (simpler orientation, better zeropadding all around), I would download it and then run this:
3dcopy T2_template.nii.gz __temp
3drefit -oblique_recenter __temp+tlrc.HEAD
3drefit -deoblique __temp+tlrc.HEAD
3dresample -orient RAI -prefix T2_template_RAI0.nii.gz -input __temp+tlrc.HEAD
3dZeropad \
-prefix T2_template_RAI0_ZP.nii.gz \
-S -100 \
-I -65 \
-L -80 \
-R -80 \
-A 10 \
-P 15 \
T2_template_RAI0.nii.gz
\rm __temp*
… which produces a dset that looks like:
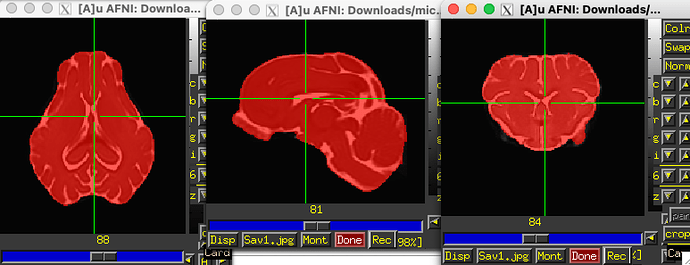
A second unfortunate thing is that there isn’t a brainmask for this dataset, and the values in the T2 template don’t stop sharply—there are small values outside where the brain is apparent. So, to create a mask, I ran:
3dcalc \
-overwrite \
-a T2_template_RAI0_ZP.nii.gz \
-expr 'step(a-10)' \
-prefix _tmp_mask0.nii.gz
3dmask_tool \
-overwrite \
-dilate_input 1 -2 3 -2 \
-prefix T2_template_RAI0_ZP_mask.nii.gz \
-input _tmp_mask0.nii.gz
\rm _tmp*
… and the mask looks like this, when overlaid on the T2 template:
OK, things are ready to go, then, perhaps. Note, because we only have a T2w image as reference, you will have to use a cost function that can match datasets with differing tissue contrasts (because you have a T1 volume). That is OK and doable in AFNI. So, your final @animal_warper command could look like:
@animal_warper \
-input DSET_ANAT \
-input_abbrev subj_anat \
-base T2_template_RAI0_ZP.nii.gz \
-base_abbrev tmplt \
-brainmask T2_template_RAI0_ZP_mask.nii.gz \
-cost lpc+ZZ \
-outdir odir_aw \
-ok_to_exist
The lpc and related lpc+ZZ cost function were developed by Saad et al. 2009 to align EPI and T1w anatomicals, and that is similar to the job here. The “+ZZ” makes the program run slower, but it has some extra stabilization for the cost estimate.
Sooo, there is some extra work involved to get the template, and then fix/add some things to it. But please let me know how that goes.
–pt
Just to add on to Paul’s response while I think of it…
There are several varieties of pigs that have templates and atlases available for them, so you may want to consider these alternatives - Yucatan micropig, Yucatan minipig, Gottingen minipig and the domestic pig.
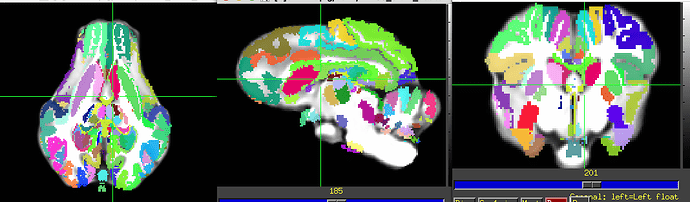
I downloaded this template and atlas from the github site for the Benn PN-150 pig brain available here:
[GitHub - neurabenn/pig_connectivity_bp_preprint: Associated files and code with the pig WM atlas and connectivity blueprint]
based on this preprint:
[https://www.biorxiv.org/content/10.1101/2020.10.13.337436v4]
That data is oblique, in the wrong orientation and not labeled properly. It’s fixable though with this small script.
3dWarp -deoblique -prefix PN150_brain_deob.nii.gz PNI50_brain.nii.gz
3dWarp -deoblique -prefix pig_saikali_deob.nii.gz -overwrite -NN pig_saikali.nii.gz
3dWarp -deoblique -prefix PNI50_MaxProb_deob.nii.gz -overwrite -NN PNI50_MaxProb.nii.gz
3drefit -orient RIP -deoblique *_deob.nii.gz
3drefit -space pig_PNI50 *_deob.nii.gz
3drefit -cmap INT_CMAP pig_saikali_deob.nii.gz PNI50_MaxProb_deob.nii.gz
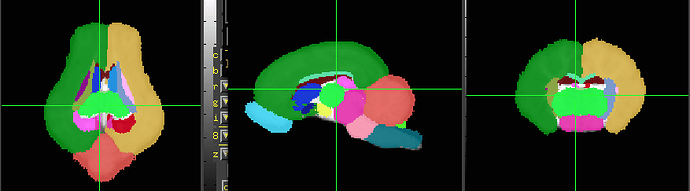
Saikali dataset over PNI50 T1 template.
They seem to use the Saikali template from here. They should have their own atlas too, but it may be only upon request.
The Illinois 4-wk and 12-wk pig atlases are available here:
[Download the Pig Brain Atlas]
The data is also oblique, so it requires similar processing.
3dcalc -a Regions_of_Interest_4wk/ROI_thr25/Cerebellum_thr25.nii -expr 0 -prefix zero.nii.gz
3dTstat -argmax -prefix MPM_4wk.nii.gz "zero.nii.gz Regions_of_Interest_4wk/ROI_thr25/*.nii"
3drefit -space ILL_PIG_4wk Pig*_4wk.nii MPM_4wk.nii.gz
3dWarp -deoblique -prefix Pig_Brain_Atlas_4wk_deob.nii.gz Pig_Brain_Atlas_4wk.nii
3dWarp -deoblique -prefix MPM_4wk_deob.nii.gz -NN -overwrite MPM_4wk.nii.gz
3drefit -cmap INT_CMAP MPM_4wk_deob.nii.gz
the Yucatan minipig
[VT Yucatan Brain Template]
[https://www.sciencedirect.com/science/article/pii/S1053811921002925]
The Gottingen minipig (another variety of pig) template, but you may need to sign an agreement with this group to get their data.
[https://www.sciencedirect.com/science/article/pii/S1053811901909103]
Hi
I need another help from you.
The thing I have done before did not work.
I have created a mask from T2 and resampled it on T1 but as I can see now it was a mess since its involves brain non brain tissue and failed to register with an atlas.
Now I have to create mask again from T1 but also since different pig head was in different position during the MRI, I can not even use the same mask for all. It would be too time consuming. Do you have any ideas how to do it. I have been googling and i saw machine or deep learning is another method but i have no prior knowledge.
Another problem is that drawing the mask seems too hard as T1 does not have that high resoultion and it is too hard for identifying the brain and non brain tissue.
Since registration is a important step. without doing the skull stripping correctly it would fail all other steps.
Thank you for your time,
Best,
Sanjida